Fig. 1.
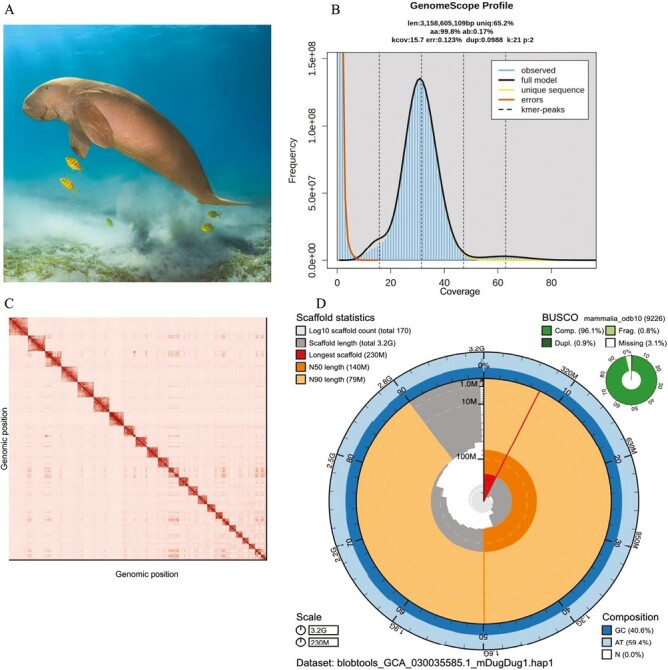
Dugong high-quality reference assembly. A) An adult dugong. Margarita Granovskaya via stock.adobe.com B) K-mer spectrum output generated from adapter-filtered PacBio HiFi data using GenomeScope 2.0. The bimodal pattern observed corresponds to a diploid genome. K-mers covered at lower coverage and lower frequency correspond to differences between haplotypes, and the higher coverage and higher frequency k-mers correspond to the similarities between haplotypes. C) Omni-C Contact maps for the curated genome assembly of haplotype 1 generated with PretextSnapshot. Omni-C contact maps translate proximity of genomic regions in 3D space to contiguous linear organization. Each cell in the contact map corresponds to sequencing data supporting the linkage (or join) between two of such regions. Scaffolds are separated by black lines. D) BlobToolKit Snail plot showing a graphical (continued next page) representation of the quality metrics presented in Table 2 for the Dugong dugon assembly for haplotype 1 (mDugdug1.hap1). The plot circle represents the full size of the assembly. From the inside out, the central plot covers length-related metrics. The red line represents the size of the longest scaffold; all other scaffolds are arranged in size order moving clockwise around the plot and drawn in gray starting from the outside of the central plot. Dark and light orange arcs show the scaffold N50 and scaffold N90 values. The central light gray spiral shows the cumulative scaffold count with a white line at each order of magnitude. White regions in this area reflect the proportion of Ns in the assembly. The dark versus light blue area around it shows mean, maximum, and minimum GC versus AT content at 0.1% intervals. The legend in the lower left indicates the scale of the circumference (3.2G total assembly size) and radius (230M longest scaffold) of the main plot. A summary of complete, fragmented, duplicated, and missing BUSCO genes in the mammalia_odb10 set is shown in the top right (Challis et al. 2020).
