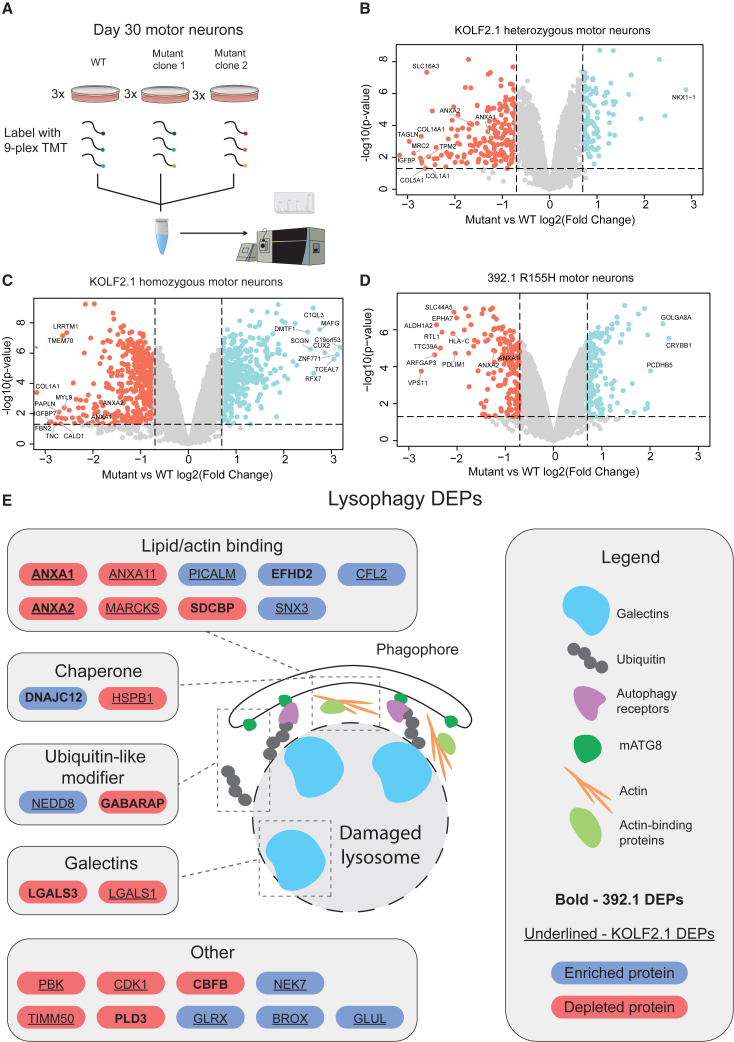
Figure 3.
Quantitative proteomics identifies autophagy defects in p97 R155H motor neurons
(A) Schematic of the experimental setup for the KOLF2.1 cells. Three replicates each of wild type (WT) and two clones of heterozygous or homozygous p97 R155H motor neurons were used in two 9-plex TMT experiments. Three replicates of WT and p97 R155H 392.1 motor neurons were used for a 6-plex TMT experiment.
(B–D) Volcano plots for the indicated cell line. Cutoff values of 0.7 log2 fold change and 0.05 p value were used to determine differentially expressed proteins (DEPs).
(E) Lysophagy-related proteins that were significantly altered in R155H motor neurons. Red and blue backgrounds indicate depleted and enriched proteins, respectively. See also Figures S4 and S5.