Figure 3.
A distinct PDGFRAlow subset retained the osteochondral competency of MSX1+ mesenchymal progenitors
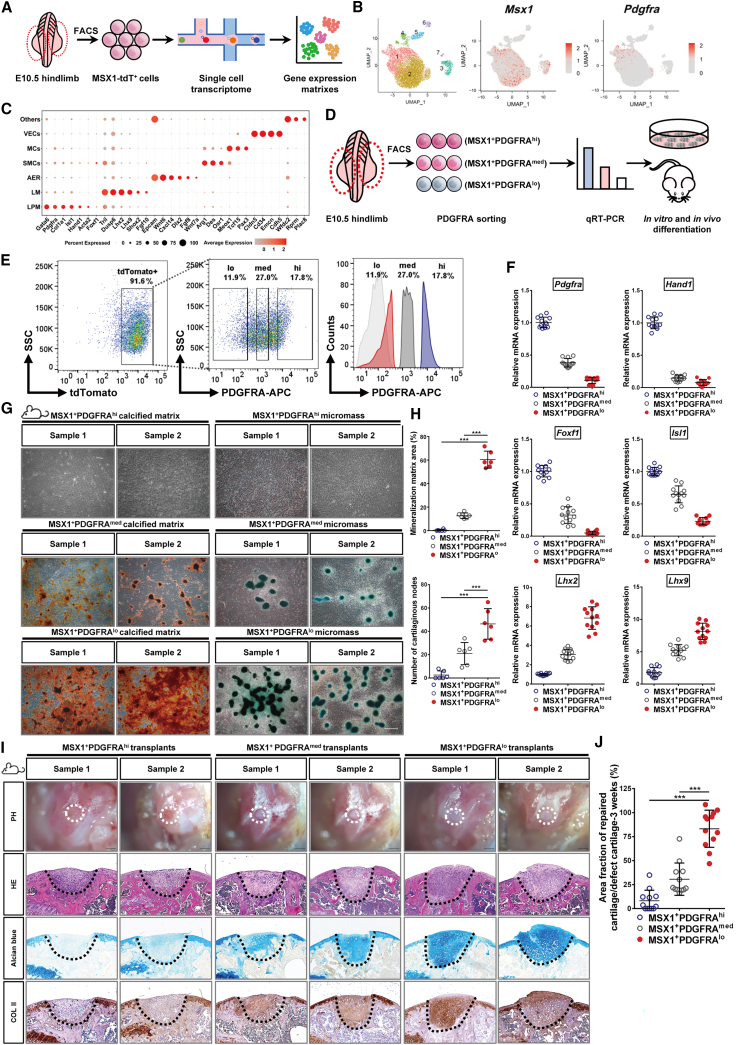
(A) Schematic diagram showing scRNA-seq analysis of E10.5 limb bud MSX1+ (tdTomato+) mesenchymal progenitor cells.
(B) Seven cell clusters were identified from scRNA-seq analysis (UMAP plots). Expression of Msx1 and Pdgfra are shown.
(C) Specific marker expression of different cell clusters. Cluster 1: lateral plate mesoderm (LPM); cluster 2: limb mesenchyme (LM); cluster 3: apical ectodermal ridge (AER).
(D) Schematic diagram of MSX1+ cell sorting and characterization strategy.
(E) Isolation of MSX1+ subpopulations by cell sorting. Left: flow cytometry analysis of MSX1+ cells from E10.5 embryos. Middle and right: gating strategy to isolate PDGFRA-expressing cells.
(F) qRT-PCR analysis of marker gene expressions for LPM (Foxf1, Hand1, Pdgfra, and Isl1) and LM (Lhx2 and Lhx9) in sorted cells. Error bars represent data from four independent experiments with triplicates.
(G) Confirmation of the osteochondral competency of MSX1+PDGFRAlow subpopulation in vitro. Differentiated cells from the sorted subsets were stained with alizarin red for osteogenic (left) and Alcian blue for chondrogenic lineages (right). Scale bars: 100 μm.
(H) Quantitative analysis of the differentiated mineralized matrix and cartilaginous nodes by sorted cells. MSX1+PDGFRAlow cells formed the largest area of the mineralized matrix and the highest number of cartilage nodes. Error bars represent data from six samples of three independent experiments (mean ± SD). Statistics: one-way ANOVA followed by Tamhane’s T2 (mineralized matrix) and Tukey (cartilaginous nodes) post hoc multiple comparisons using SPSS version 22.0. ∗∗∗p < 0.001.
(I) MSX1+PDGFRAlow cells could efficiently regenerate articular cartilage in vivo. Representative images of 3 week samples are shown. Alcian blue and COL II staining (black dashed line) were used to detect cartilage formation. Scale bars: 1 mm (BF) and 200 μm (H&E, Alcian blue, and COL II).
(J) Quantitative analysis of cartilage repair in the defect sites at 3 weeks. Error bars represent data from twelve sections of six mice in three independent experiments (mean ± SD). Statistics: one-way ANOVA followed by Tukey post hoc multiple comparisons using SPSS version 22.0. ∗∗∗p < 0.001.