Figure 1.
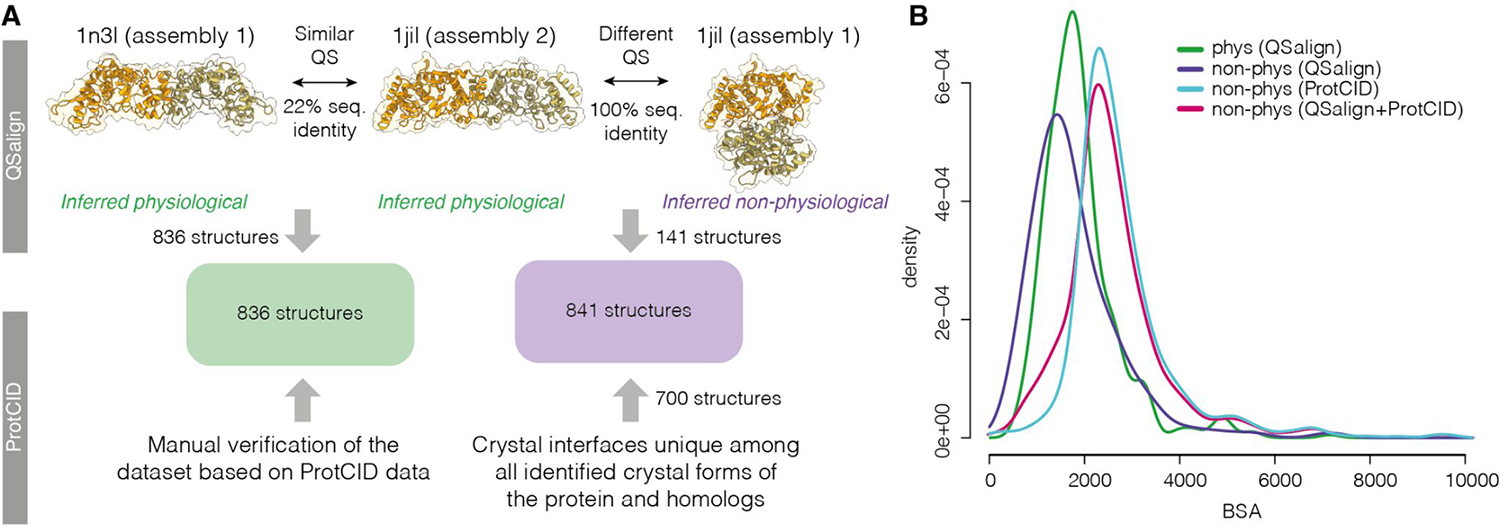
Approach used to define the benchmark dataset. A. Physiological dimers (836 in total) were identified based on the QSalign resource [34] and have their interface structure conserved across homologues (e.g., 1n3i vs 1jil). A manual curation of that set was also performed based on the ProtCID resource [33]. A subset of non-physiological dimers (141 structures) were identified based on QSalign as structures exhibiting a different and conserved interface. For example, two assemblies are available in the PDB for structure 1jil. Assembly 2 shows a conserved interface, which lets us infer that Assembly 1 is non-physiological. This set was expanded to also include dimers forming interfaces that are unique among all interfaces across crystal forms or across the crystal forms of homologs, as defined in ProtCID, yielding an additional 700 structures. B. Interfaces area distributions of the physiological and non-physiological homodimers and the sets described in panel A.
