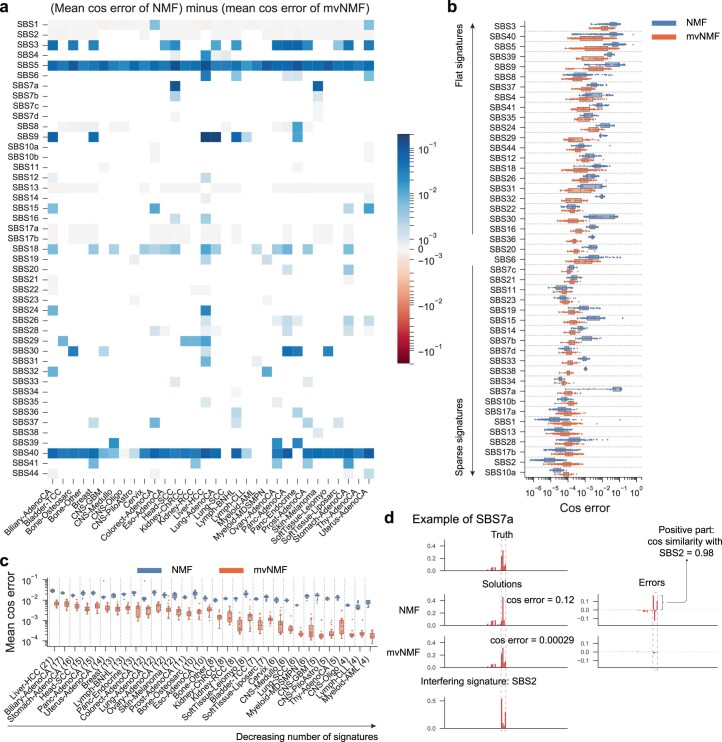
Extended Data Fig. 1. mvNMF improves the accuracy of de novo signature discovery in simulated datasets with SBS signatures.
Synthetic datasets are simulated from tumor type-specific SBS signatures and Dirichlet-distributed exposures for 32 tumor types with 20 replicates. Each dataset contains 200 samples and on average 5,000 mutations per sample (Poisson-distributed). NMF and mvNMF are then applied for de novo signature discovery, assuming that the number of signatures is known. Finally, the discovered signatures are compared to the true ones, and their discrepancies are quantified by cosine errors. a. Heatmap of the difference between NMF- and mvNMF-derived cosine errors. Each element represents the mean of 20 independent simulations. b. NMF- and mvNMF-derived cosine errors for different SBS signatures, sorted by standard deviation of the corresponding signature spectrum. Data from different tumor types are collapsed. Same as Fig. 2c and included for completeness. c. NMF- and mvNMF-derived cosine errors for different tumor types, sorted by the number of signatures present in the corresponding tumor type. Data from different signatures are averaged within each tumor type. n = 20 independent simulations for each box plot. d. An example comparing the performance of NMF and mvNMF on identifying SBS7a. The NMF solution of SBS7a receives a large cosine error. The error spectrum indicates interference from SBS2 coexisting in the dataset. By comparison, mvNMF does not suffer from the SBS2 interference and is able to discover SBS7a accurately.