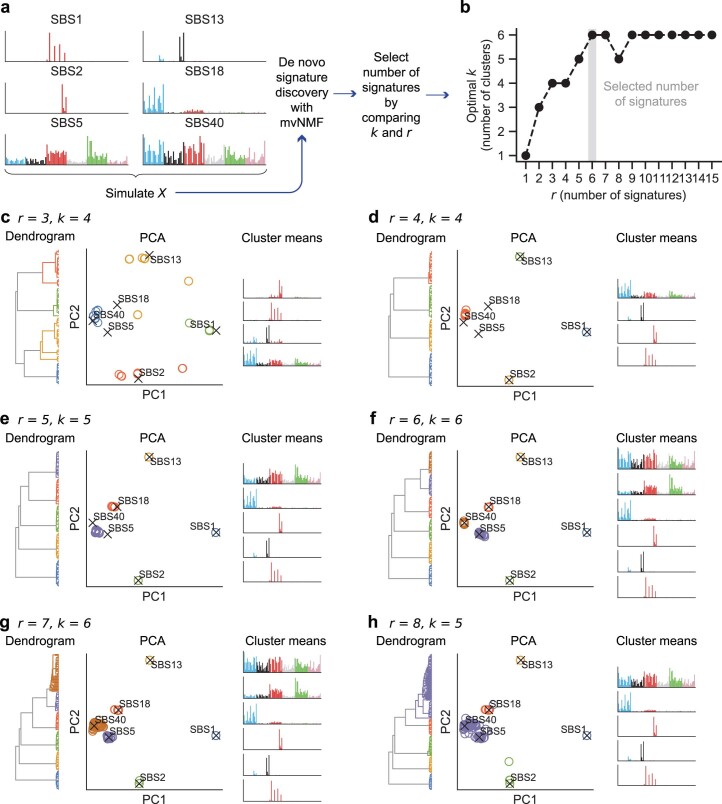
Extended Data Fig. 2. An example illustrating the method for selecting the number of signatures r.
a. Data simulation scheme. A synthetic dataset is simulated from Dirichlet-distributed exposures and 6 SBS signatures (SBS1, 2, 5, 13, 18, and 40) believed to be present in cervical cancers according to PCAWG results. The dataset contains 200 samples and on average 5000 mutations per sample. mvNMF is then run for r = 1 to 15 separately, with 20 independent replicates from different initializations for each r value. b. Selecting the number of signatures by comparing k and r. De novo signatures from solutions with the same r are clustered into k clusters, where k is determined by the gap statistic60,61. The greatest r such that k = r is chosen as the final number of signatures. In this example, the correct number of signatures (r = 6) is selected. c-h. Details of mvNMF solutions at different r values. For each r value, the mvNMF-derived de novo signatures from 20 independent runs are clustered and visualized as dendrograms and PCA plots. The signatures corresponding to cluster means are also shown. When r is smaller than the true number of signatures (for example, r = 3), the mvNMF solutions can be unstable, resulting in k > r. When r is greater than the true number of signatures (for example, r = 7), mvNMF may produce redundant signatures, resulting in k < r. Together, these observations suggest that the greatest r with k = r is a reasonable estimate of the true number of signatures.