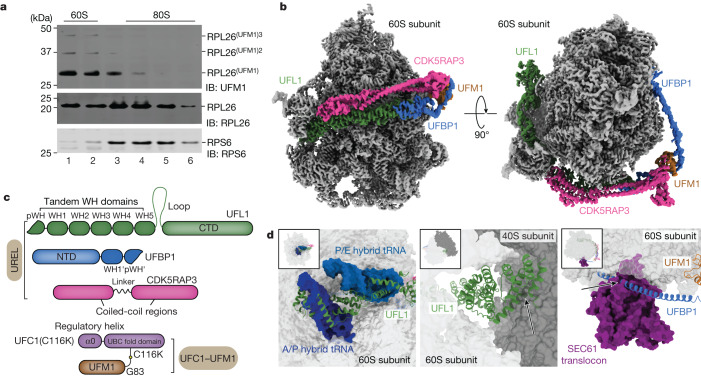
Fig. 1. Cryo-EM structure of the 60S–UREL–UFC1–UFM1 complex.
a, ER-associated 60S ribosomal subunits are preferentially UFMylated in cells. The membrane fractions from WT HEK293 cells were solubilized and layered over a 10–30% sucrose gradient to separate individual ribosomal subunits. The 60S and 80S fractions were analysed for RPL26 UFMylation by immunoblotting (IB) using the indicated antibodies. The blot is representative of n = 3 independent experiments. Superscript values 2 and 3 refer to di- and tri-UFM1 RPL26 modifications, respectively. b, Composite cryo-EM density of the UREL ligase complex and UFM1 bound to the 60S ribosome. The 60S ribosome map is coloured in grey, and the UREL and UFM1 density map is coloured by protein as shown in c. c, Schematic of the domain architecture of UREL components (UFL1, UFBP1, CDK5RAP3) and the UFC1–UFM1 mimic. WH, winged-helix domain. NTD, N-terminal domain. d, Superpositions of UREL with published ribosome structures highlighting clashes with 60S ribosome-binding components. Left, the UFL1 C-terminal domain (CTD) occupies A-, P- and E-tRNA-binding sites and clashes with superimposed A/P and P/E tRNA (Protein Data Bank (PDB): 6W6L). Middle, the UFL1 CTD clashes with the 40S subunit (PDB: 6ip8). Right, the UFBP1 helix clashes with the SEC61 translocon (PDB: 6r7q). The arrows indicate clashes.