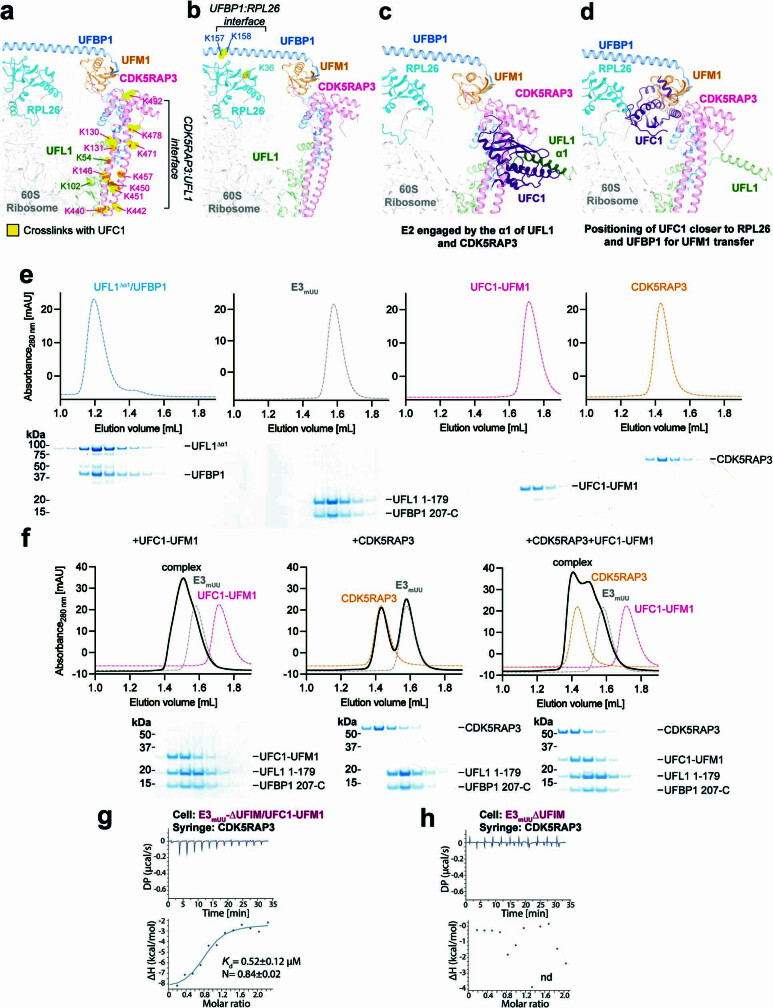
Extended Data Fig. 7. Existence of a composite binding site for UFC1~UFM1 on UREL.
a, Mapping crosslinked residues on the UREL:60S cryo-EM structure. Residues on CDK5RAP3 and UFL1 crosslinked with UFC1 are highlighted in yellow. b, Residues on UFBP1 and RPL26 crosslinked with UFC1 may constitute two distinct interfaces for charged E2 interaction. Cryo-EM model of UREL:60S is shown in cartoon representation and crosslinked residues shown as ball and sticks are highlighted in yellow. (c-f) Models depicting different intermediate stages of UFC1 prior to conjugation of UFM1 on RPL26. c, Model to show that UFC1 is potentially engaged closer to the interface formed by CDK5RAP3 and UFL1 as observed in XL-MS data shown in (a). Model was generated by superposition of crystal structures of E3mUU bound to UFC1 and cryo-EM structure of UREL:60S ribosome. d, Model generated by superposition of crystal structure of UFC1-UFM1 conjugate onto cryo-EM structure of UREL:60S ribosome to suggest UFC1’s proximity to RPL26 and UFBP1 as observed in the XL-MS data shown in (b). e, (Top) Individual SEC elution profiles for UFL1-Δα1/UFBP1, E3mUU, CDK5RAP3 and UFC1-UFM1. (Bottom) The corresponding peak fractions were separated on a 4-12% SDS-PAGE gel under reducing conditions and Coomassie stained. f, (Top) Gel filtration chromatograms of E3mUU:UFC1-UFM1, E3mUU:CDK5RAP3 and E3mUU:CDK5RAP3/UFC1-UFM1. (Bottom) The corresponding peak fractions were separated on a 4-12% SDS-PAGE gel under reducing conditions and Coomassie stained. g, ITC binding curves for preformed E3mUU/UFC1-UFM1 complex and CDK5RAP3. Data are representative of n = 2 independent experiments. The dissociation constant and stoichiometry were calculated based on both experiments. h, Control experiment for (g), where no UFC1-UFM1 was added to E3mUU. nd indicates that no binding was detected. Data are representative of n = 2 independent experiments.