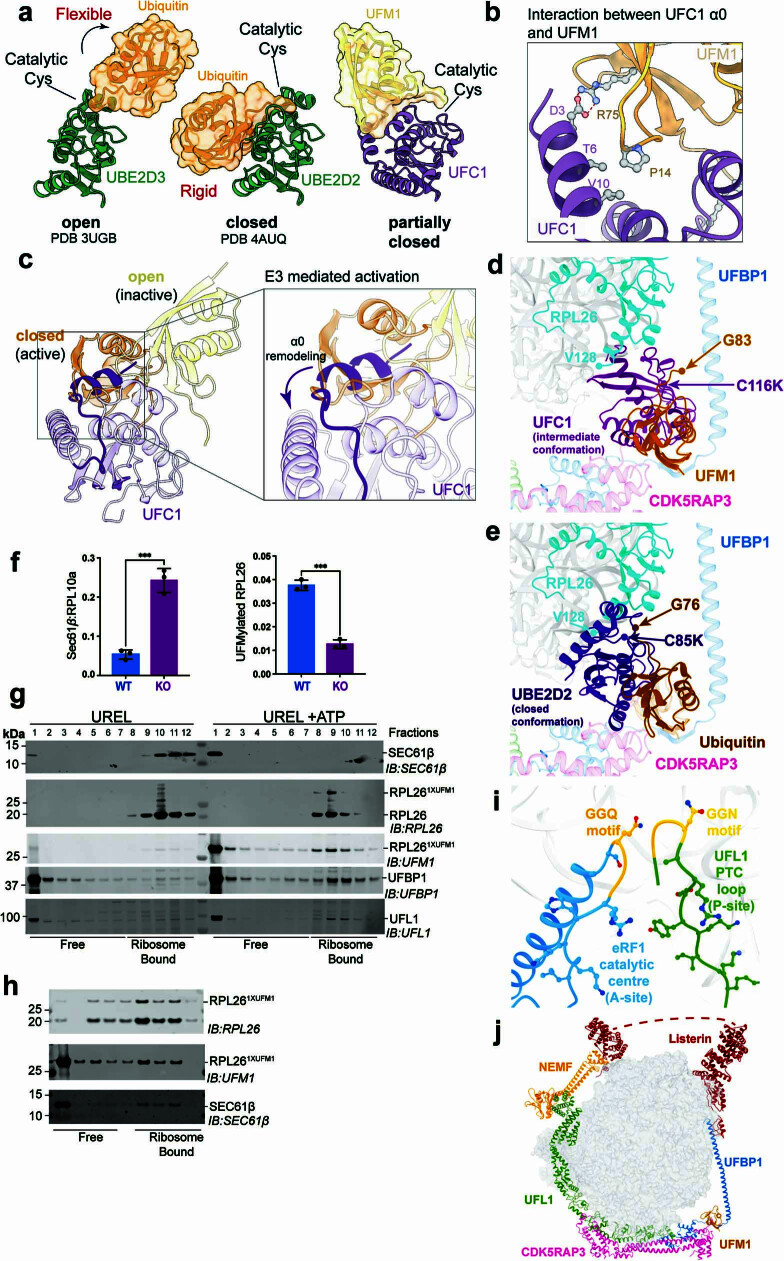
Extended Data Fig. 8. Active conformation of UFC1~UFM1 and UFMylation-dependent SEC61 dissociation from 60S.
a, Comparison of apo and E3 bound states of E2~Ubiquitin conjugate (PDB IDs 3ugb, 4auq) with apo UFC1-UFM1 conjugate. Ubiquitin (dark orange) and UFM1 (light orange) are shown as surfaces overlaid on cartoons. UBE2D2/3 (green) and UFC1 (purple) are shown in cartoon representation. b, Enlarged view of the interface between UFC1 α0 and UFM1 with the interacting residues highlighted in ball and stick representation. c, Model depicting open and closed states of UFC1~UFM1 conjugate. Inset highlights the clash between UFC1 α0 and an incoming UFM1 suggesting a requirement to remodel UFC1 α0 to accommodate UFM1 in the closed-active state. d, Crystal structure of UFC1-UFM1 conjugate in an intermediate conformation superimposed onto cryo-EM structure of UREL:UFM1 bound 60S ribosome. C-terminal glycine of UFM1 (G83), catalytic cysteine to lysine mutation of UFC1 (C116K) and most C-terminal residue of RPL26 for which density is present in the cryo-EM map (V128) are depicted as circles. e, Crystal structure of Ubiquitin-E2 conjugate (PDB ID 7r71) in a closed conformation superimposed onto the cryo-EM structure of UREL:UFM1-bound 60S ribosome in the same view as (d). C-terminal glycine of ubiquitin (G76), catalytic cysteine to lysine mutation C85K of ubiquitin E2 (UBE2D2) and most C-terminal residue of RPL26 for which density is present in the cryo-EM map (V128) are depicted as circles. f, Quantification of immunoblots from (Figure 5b). UFMylated RPL26 band intensity normalized to intensity of total RPL26 (left) and SEC61β band intensities normalized to RPL10a. Data show mean ± SD. ***p < 0.0001 (Student t-test). Data are representative of n = 2 independent experiments. g, UFMylation mediates dissociation of 60S from the translocon. In vitro UFMylation reactions were performed on membrane associated 60S ribosomal subunit-SEC61 translocon complexes isolated from CDK5RAP3 KO cells. 60S-SEC61 complexes were incubated with UBA5, UFC1, UREL and UFM1 either in the presence or absence of ATP and the reaction products were separated on a sucrose gradient and analysed by immunoblotting with the indicated antibodies. Blot is representative of n = 2 independent experiments. h, Dissociation of 60S from the ER translocon requires the UFIM motif. In vitro 60S-SEC61 UFMylation and translocon dissociation assay performed as in (d) using UREL complex with the UFBP1 UFIM mutant F196A. Blot is representative of n = 2 independent experiments. i, Comparison of the UFL1 PTC loop (P-site) with the eRF1 catalytic centre (A-site; PDB ID 6ip8). eRF1 catalytic residues GGQ and UFL1 loop residues GGN coloured in yellow. j, Superimposition of UREL complex and NEMF:Listerin complex (PDB ID 3j92) bound to the 60S ribosome. Missing Listerin model is depicted as dashed line.