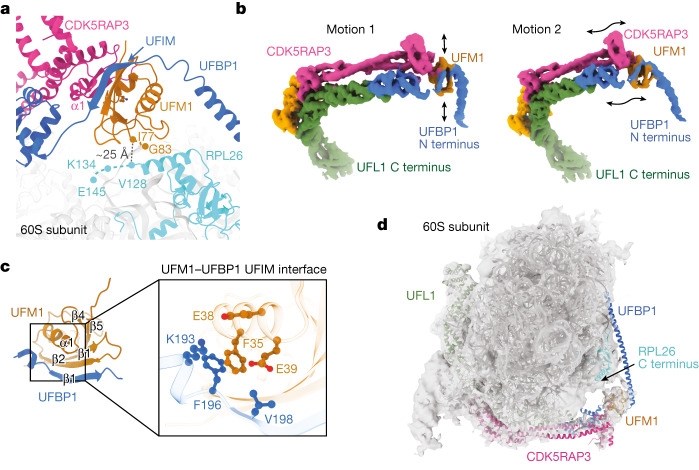
Fig. 3. The UFIM motif of UFBP1 binds to UFM1.
a, View of the catalytic region of the UREL complex bound to the 60S ribosome showing the UFBP1 UFIM and CDK5RAP3 N terminus bound to UFM1. The distance between the C termini of UFM1 and RPL26 in our cryo-EM model is shown as a black dashed line. Missing residues of UFM1 and RPL26 are shown as dashed lines in their respective colours. b, 3DFlex was used to generate motion models of the UREL ligase complex. The black arrows indicate the direction of motion observed. c, AlphaFold prediction of the UFBP1 UFIM–UFM1 complex. Inset: an enlarged view of the UFBP1 β1 and UFM1 β2 interface; key interactions are highlighted. d, Cryo-EM density map of UFMylated 60S ribosomes shown in transparent grey with a model of UREL ligase bound to 60S rigid-body fitted into the density.