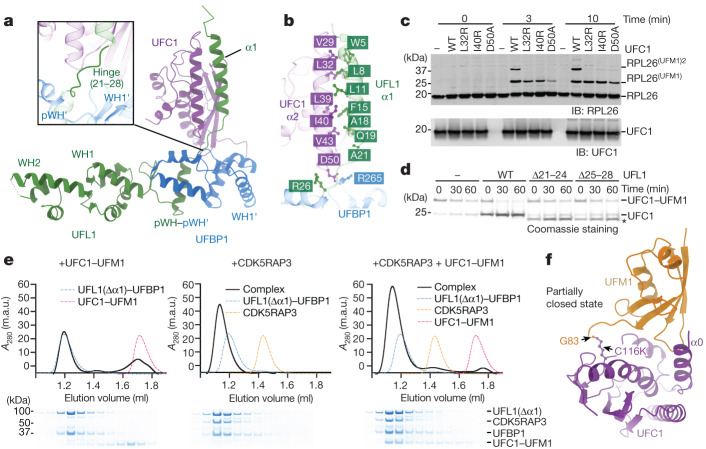
Fig. 4. The mechanism of E2 recognition and UFMylation.
a, The crystal structure of E3mUU(ΔUFIM) bound to UFC1 shown as a cartoon representation. Inset: enlarged view of the hinge region that connects UFL1 WH1 to α1. b, Enlarged view of the interaction between UFC1 α2 and UFL1 α1. c, Immunoblot analysis of in vitro 60S ribosome UFMylation in the presence of the indicated E2 mutants that are defective in E3 binding. The blot is representative of n = 3 independent experiments. d, Lysine-discharge assays in the presence of E3mUU bearing truncations in the hinge region. The asterisk indicates UFL1(Δ21–24) and UFL1(Δ25–28). The Coomassie-stained SDS–PAGE gel is representative of n = 3 independent experiments. e, CDK5RAP3 and UFL1 form a composite binding site for the UFC1–UFM1 conjugate. Top, SEC elution profiles of UFL1(Δα1)–UFBP1–UFC1–UFM1, UFL1(Δα1)–UFBP1–CDK5RAP3 and UFL1(Δα1)–UFBP1–CDK5RAP3–UFC1–UFM1 complexes. Bottom, the corresponding peak fractions were separated on a 4–12% SDS–PAGE gel under reducing conditions and Coomassie stained. A280, absorbance at 280 nm. f, The crystal structure of the UFC1–UFM1 conjugate shown as a cartoon representation. UFC1 is coloured in purple, UFM1 is coloured in orange and the isopeptide bond formed between UFM1 Gly83 and UFC1(C116K) is highlighted and shown as a ball and stick representation.