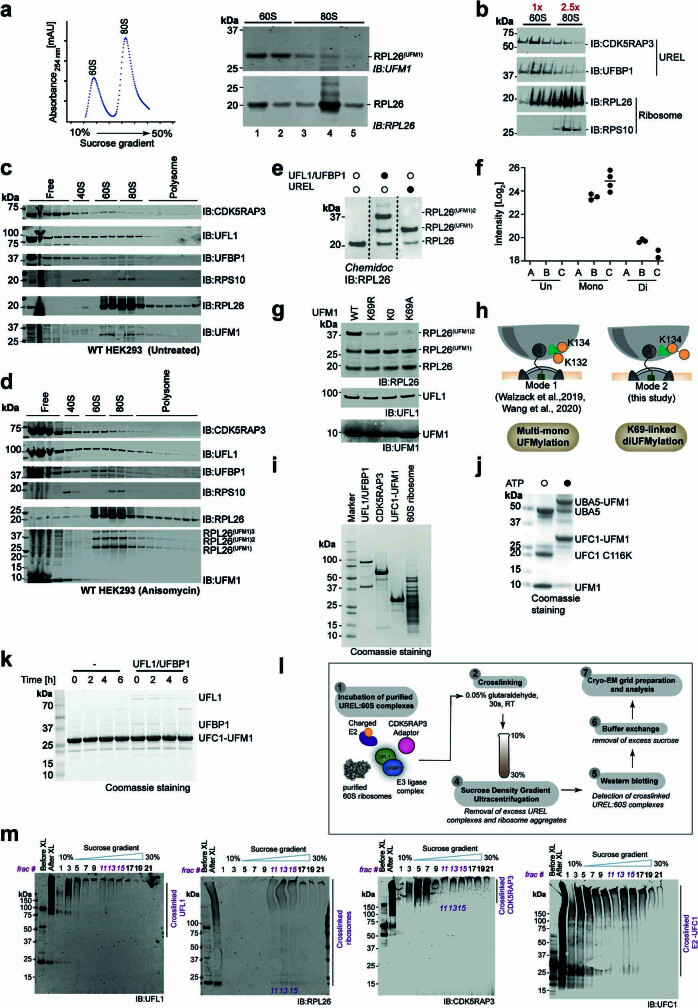
Extended Data Fig. 1. Preparation of complexes for cryo-EM.
a, UREL selectively UFMylates 60S in vitro. Ribosome UFMylation by the addition of UBA5, UFC1, UFM1 and UREL (UFL1, UFBP1 and CDK5RAP3) to a mixture of 60S ribosomes and 2-fold excess of 80S ribosomes. The reaction mixture was separated on a sucrose density gradient (left) and the fractions analysed for RPL26 UFMylation by immunoblotting (right). Data are representative of n = 3 independent experiments. b, UREL binds to 60S ribosomes. UREL was incubated with a mixture of 60S and 2.5-fold excess 80S ribosomes, separated on a sucrose density gradient and analysed by immunoblotting with the indicated antibodies. c & d, UREL associates with UFMylated 60S ribosomes in cells. Membrane fractions from HEK 293 WT cells left untreated (top) or treated with anisomycin (bottom) were separated on a sucrose density gradient and the different fractions were immunoblotted using the indicated antibodies. e, In vitro UFMylation assay to generate UFMylated 60S ribosomes in the presence of UFL1/UFBP1 or UREL for LC-MS/MS analysis. f, The reaction products from (e) were analysed by LC-MS/MS to identify the UFMylation site, linkage type and to quantify UFMylation of RPL26 under different conditions. Abundance of K134-GG remnants in the presence of E1 and E2 alone (A), UFL1/UFBP1 (B) and UREL (C). (Un: Unmodified RPL26, Mono: MonoUFMylated RPL26, Di: DiUFMylated RPL26) (n > 2 technical replicates). g, Immunoblot showing UFMylation of RPL26 in the presence UFM1 WT, K69R, K69A or lysine-less UFM1(K0). h, Mode of UFMylation of RPL26 as inferred from LC-MS/MS and biochemical experiments from e to g. i, Coomassie stained SDS-PAGE gel of purified UREL, UFC1-UFM1 and 60S ribosomes used in the preparation of samples for visualization by cryo-EM. j, Coomassie stained SDS-PAGE gel showing in vitro reaction for the generation of UFC1-UFM1. k, Coomassie stained SDS-PAGE gel analysing stability of UFC1-UFM1. Purified UFC1-UFM1 was incubated with UFL1/UFBP1 at 37 °C to monitor hydrolysis of UFC1-UFM1 conjugate. The reaction was stopped at indicated time points and separated on a 4-12% SDS-PAGE gel followed by Coomassie staining. l, Schematic showing reconstitution of stable UREL:60S complexes for cryo-EM analysis. m, Preparation of stable UREL:60S complex as outlined in l.