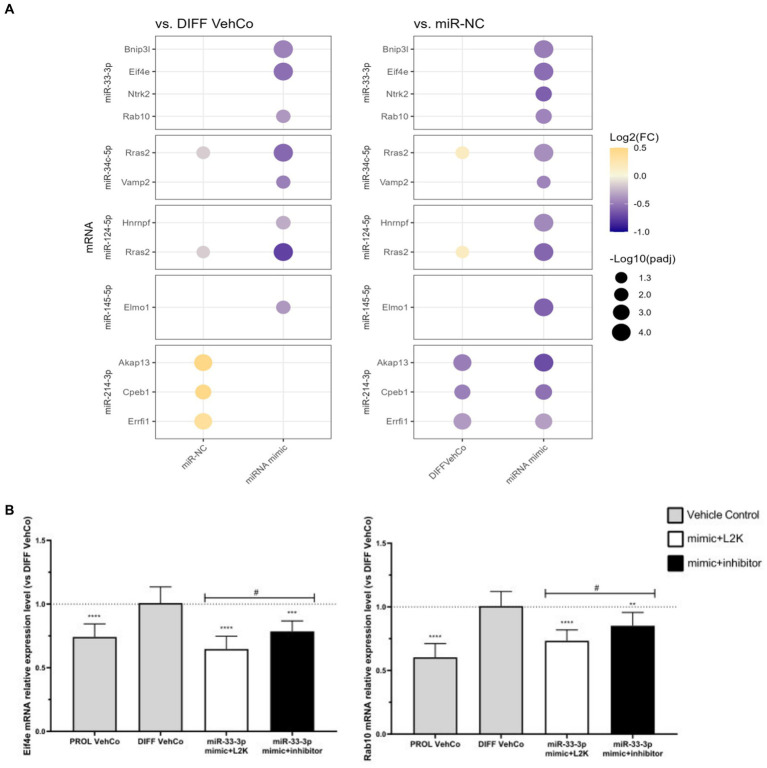
Figure 8.
Potential mRNA targets of each microRNA verified in RT-qPCR. (A) This dot plot shows the fold change (FC) of the relative expression of potential mRNA targets of each microRNA as labeled on the y-axis, to the differentiation vehicle control (DIFF VehCo) and the microRNA mimic negative control (miR-NC), resulting from the single transfection of each microRNA mimic in CG-4 cells in differentiation culture conditions. The color of each dot represents the fold change (expressed in logarithm base 2 [Log2(FC)]) of the relative expression, scaled by a blue-to-yellow color gradient from −1 to 0.5 (blue corresponding to a negative fold change, yellow to a positive), and its size represents the adjusted p-value (padj) (expressed as minus logarithm base 10 [−Log10(padj)]) as indicated in the plot legend. The adjusted p-value was calculated by Dunn’s or Dunnett’s multiple comparison test to each control separately, i.e., the differentiation vehicle control and miR-NC, as recommended following a parametric/non-parametric one-way ANOVA on all conditions, including the proliferation vehicle control (not shown). Only the significant data are depicted; thus, missing dots correspond to non-significant data. −Log10(padj) ≥1.3 corresponds to the significance level of p ≤ 0.05, −Log10(padj) of 2.0 corresponds to p = 0.01, −Log10(padj) of 3.0 to p = 0.001, and −Log10(padj) of 4.0 to p = 0.0001. Data were from three experiments with each condition in triplicate. The corresponding bar plots showing both the statistically significant and non-significant results can be found in Supplementary Figure S5A. (B) The plots show the relative expression (mean with SD of 2−ΔΔCt) of Eif4e and Rab10 to the differentiation vehicle control (DIFF VehCo, gray bar), resulting from the sequential transfection of miR-33-3p mimic followed (black bar) or not (white bar) by its inhibitor (as labeled on the x-axis) in CG-4 cells in differentiation culture conditions, as well as the proliferation vehicle control (PROL VehCo, gray bar). The dotted line represents the mean relative expression of the differentiation vehicle control to which the relative expression (2−ΔΔCt) of each sample was calculated. The adjusted p-value was calculated by Dunnett’s multiple comparison test to the differentiation vehicle control (*) and by Sidak’s multiple comparison test between the microRNA mimic and its inhibitor (#), as recommended following a parametric one-way ANOVA on all conditions: */# for p ≤ 0.05, ** for p ≤ 0.01, *** for p ≤ 0.001, **** for p ≤ 0.0001. Data were from three experiments with each condition in triplicate. L2K, Lipofectamine™ 2000; SD, standard deviation.