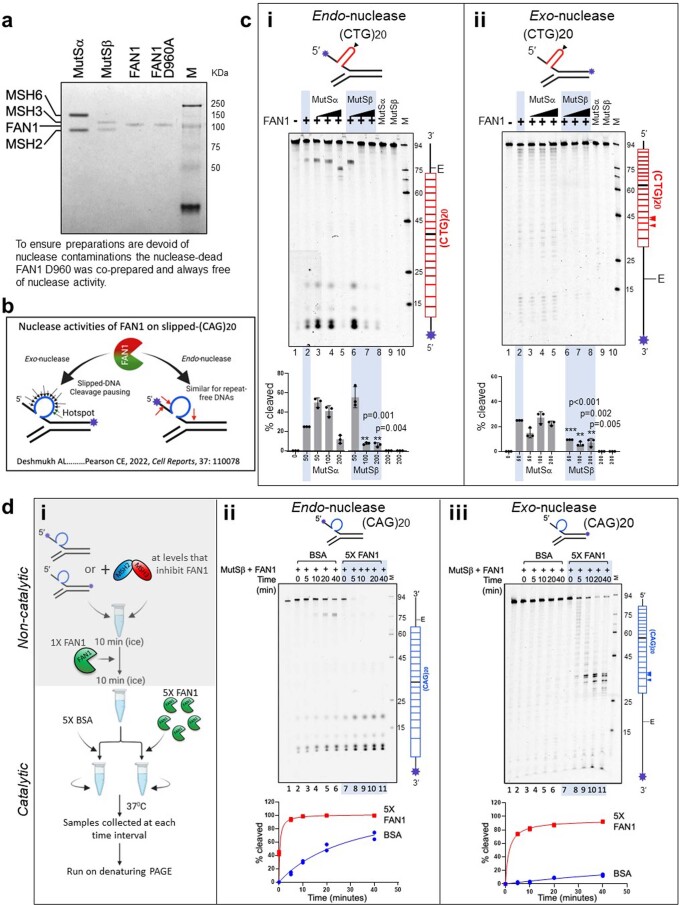
Extended Data Fig. 5. MSH2, MSH3, MSH6, and FAN1 proteins, nuclease assay, (CTG)20 digestion.
a, A representative Coomassie brilliant blue-stained SDS-PAGE of purified human MutSα (MSH2-MSH6), MutSβ (MSH2-MSH3), FAN1 and nuclease-dead FAN1p.D960A (purified in parallel to ensure an absence of contaminating nuclease activity in preparations), expressed in Sf9 cells using baculoviral overexpression. Both MutSα and MutSβ preparations were free of nuclease contamination (see c panels i and ii, lanes 9 and 10). Purity and activity were assessed for ~10 preparations, with consistent results. b, Schematic of FAN1 exo-nucleolytic and endo-nucleolytic cleavage sites on slipped-(CAG)20 DNA substrates labeled at the 3’ or 5’ ends, respectively. FAN1’s exo-nucleolytic ‘nibbling’ of excess repeats parallels the ‘inchworm’ expansions in patient brains, suggesting a role for FAN1 in regulating repeat instability46. Endo- and exo-nucleolytic activities can be distinguished by fluorescein amidite (FAM)-labeling at 5’ or 3’ ends of the (CAG)20 strand, respectively (asterisk); in this manner, only the labeled strand and its digestion products are tracked. c, Slipped-(CTG)20 DNAs are digested by endo- and exo-nucleolytic activities of FAN1, which can be inhibited by MutSβ, but not by MutSα. FAM-labelled CTG slip-out oligonucleotides (schematic, arrowhead indicated center of repeat tract) mimic putative intermediates of expansion mutations. Exo- and endo-nuclease activities were determined by labeling either 3′- or 5′-end of (CTG)20 strand, respectively (asterisk). ‘E’ is elbow at the dsDNA-ssDNA junction. Slip-out DNA substrates (100 nM) were preincubated with increasing amounts of purified human MutSα or MutSβ (0-200 nM), and nuclease digestion was initiated by addition purified human FAN1 (50 nM). Vertical schematic to the right of each gel indicates location of cleavage sites along the FAM-labeled DNA strand. Percentage cleavage (densitometry) for each reaction was then normalized to cleavage levels for FAN1 alone, and these levels were graphed (n = 3 experiments, mean ± s.d. plotted). Results of two-sided unpaired t-test are indicated (vs. ‘FAN1 alone’ in panels i and ii). d, Excess FAN1 overcomes MutSβ-mediated inhibition. (i) Flowchart outlining the competition experiment. Human MutSβ (200 nM), FAN1 (50 nM) and (ii) 5’- or (iii) 3′-FAM-labeled CAG slip-outs (100 nM), that is conditions established to be completely inhibited by MutSβ, were preincubated in non-catalytic conditions (on ice). Reactions were split at the 10-min time point and challenged with 5× BSA, to control for macromolecular crowding (lanes 2–6) or an excess (5×) of FAN1 (lanes 7–11). Aliquots were taken for analysis at different timepoints (0, 5, 10, 20 and 40 minutes) and cleavage products were quantified (n = 2 experiments).