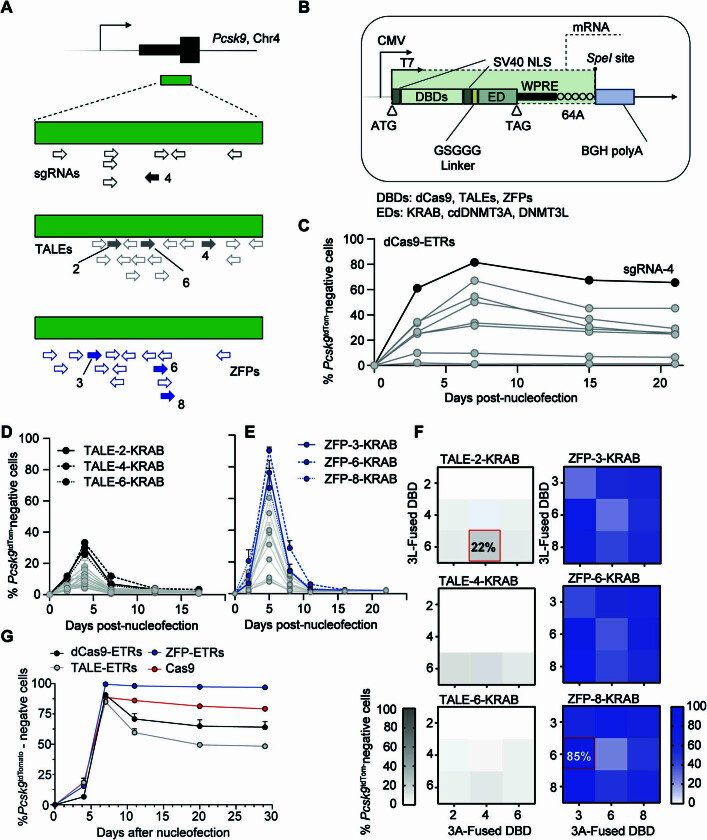
Extended Data Fig. 1. In vitro selection of the most effective ETRs for Pcsk9 silencing.
a, Schematic drawing showing on top the Pcsk9 promoter region with the annotated CpG Island (CGI) and, on the bottom, a zoom on the CGI showing the target sites of all the tested single guide RNAs (sgRNAs; black arrows), TALEs (grey arrows), and ZFPs (blue arrows). Filled arrows indicate the most active sgRNA/DBDs used for subsequent experiments. Created with BioRender.com. b, Schematic representation of the plasmid used for ETR expression, either after its direct transfection into cells or as a template for In Vitro Transcription (IVT) of the ETRs’ mRNA. CMV: enhancer/promoter of the Cytomegalovirus. T7: promoter for mRNA production. ATG: start codon; DBDs: DNA-binding domains; SV40 NLS: nuclear localization signal of the simian virus 40; GSGGG: glycine-rich liker peptide; ED: effector domain, either KRAB from the ZNF10 protein, cdDNMT3A or DNMT3L; WPRE: woodchuck hepatitis virus post-transcriptional regulatory element; 64A: stretch of 64 adenines; SpeI: restriction site used to linearize the plasmid for IVT; BGH polyA: polyadenylation signal from the bovine growth hormone gene. Created with BioRender.com. c, Dot plot showing the percentage of Pcsk9tdTomato-negative cells over a period of 22 days post-delivery of plasmids encoding for the indicated dCas9-based ETRs and 8 different sgRNAs. sgRNA-4 was the most active among the tested guides (black dots and connecting line) and thus used for subsequent experiments. Data are reported as mean (n = 2). d, Dot plot showing the percentage of Pcsk9tdTomato-negative cells over a period of 17 days post-delivery of plasmids encoding for 16 different TALE DBDs fused to the KRAB domain. This experiment was meant to identify the most effective TALEs among those tested, using KRAB-mediated epi-silencing of Pcsk9 as a surrogate readout for DBD efficiency. TALE-2, -4 and -6 were the most active ones among those tested (black dots and connecting line) and thus used for subsequent experiments. Data are reported as mean ± s.d. (n = 4). e, Dot plot showing the percentage of Pcsk9tdTomato-negative cells over a period of 22 days post-delivery of plasmids encoding for 16 different ZFP DBDs fused to the KRAB domain. This experiment was meant to identify the most effective ZFPs among those tested, using KRAB-mediated epi-silencing of Pcsk9 as a surrogate readout for DBD efficiency. ZFP-3, -6 and -8 were the most active ones among those tested (blue dots and connecting line) and thus used for subsequent experiments. Data are reported as mean ± s.d. (n = 4). f, Left: heat maps showing the percentage of Pcsk9tdTomato-negative cells at day 7 post-delivery of combinations of plasmids encoding for KRAB-, DNMT3L and cDNMT3A-based ETRs containing TALEs. The matrixes were built by transfecting either one of the TALE-KRAB ETRs from d with all possible combinations of the three best-performing TALE DBDs (namely, 3, 6 and 8) fused to either DNMT3L (y axis) or cdDNMT3A (x axis). Given its highest performance, the triple-ETR combination containing TALE-2-KRAB, TALE-6-DNMT3L, and TALE-4-cDNMT3A was chosen for further studies. Colour intensity refers to average silencing efficiency (n = 2). Right: similar experiment as in left but performed with the three best-performing ZFPs from e. Given its highest activity, the triple-ETR combination containing ZFP-8-KRAB, ZFP-6-DNMT3L and ZFP-3-cDNMT3A was chosen for further studies. Colour intensity refers to average silencing efficiency (n = 3). Best-performing triple combinations of TALE- and ZFP-ETRs are indicated with a red square. g, Time-course analysis of Pcsk9tdTomato-negative cells from the 0.5 µg RNA treatment conditions in Fig. 1b of the main text. Data are reported as mean ± s.d.