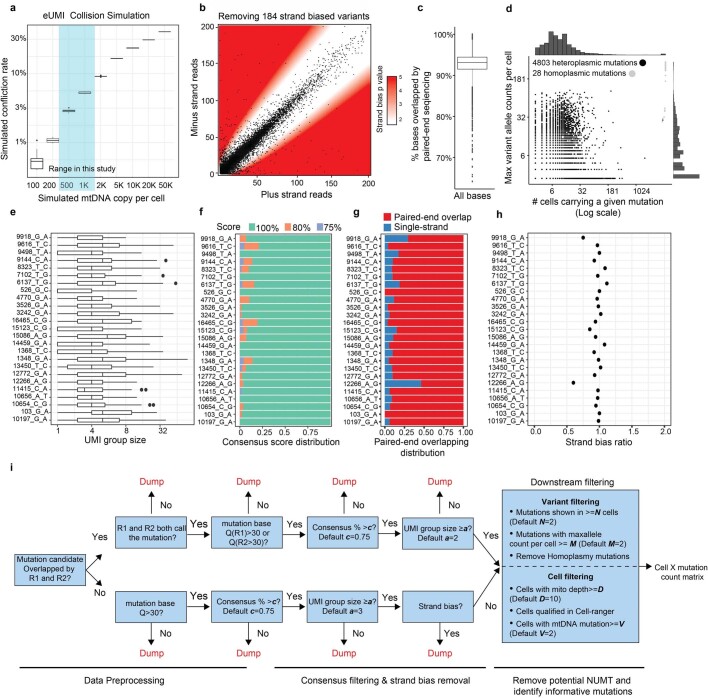
Extended Data Fig. 1. Improved mtDNA coverage and mutation detection with ReDeeM pipeline.
a, Justification of the use of endogenous UMI (eUMI) based on cell barcode plus starting and ending sites through simulation. The starting and ending sites were simulated based on empirical distribution of Tn5 cutting sites. The number of mtDNA copies per cell was simulated (x-axis) and the collision rate was calculated accordingly. n = 10 simulations in each box. Boxplot displays data from the 25th to 75th percentile, and whiskers extending to the minimum and maximum within 1.5 IQR. b-h, Quality controls for ReDeeM variant calling, related to main Fig. 1. b, Removing strand-biased mtDNA variants using a binomial distribution model. White zone includes mutations for which the null hypothesis (no strand biases) holds true, whereas red zone indicates rejection of this null hypothesis (both p < 0.01 and fold change > +/− 2) in favour of strand biases. P-values are derived from two-sided binomial test. c, Percentage of bases that were overlapped by both reads in a paired-end sequencing. n = 7,404 cells. Boxplot displays data from the 25th to 75th percentile, and whiskers extending to the minimum and maximum within 1.5 IQR. d, Summary of the mutations regarding the number of cells carrying the mutation (x-axis) and the maximum number of mutant alleles per cell (y-axis). Heteroplasmic mutations in black dots and homoplasmic mutations in grey dots. e-h, Detailed mutation quality control metrics (random examples are shown). e, The number of supporting reads for each mtDNA molecule (UMI group) containing the mutation. Boxplot displays data from the 25th to 75th percentile, and whiskers extending to the minimum and maximum within 1.5 IQR. f, Consensus score: the percentage of mutant reads in a UMI group. Mutations with consensus scores of less than 75% were discarded. g, For a given mutation, the proportion of molecules that were covered by both reads in a paired-end sequencing. h, Strand bias ratio (a ratio of 1 indicating no strand bias). i, Workflow of the ReDeeM variant calling pipeline. NUMT: Nuclear mitochondrial DNA segments.