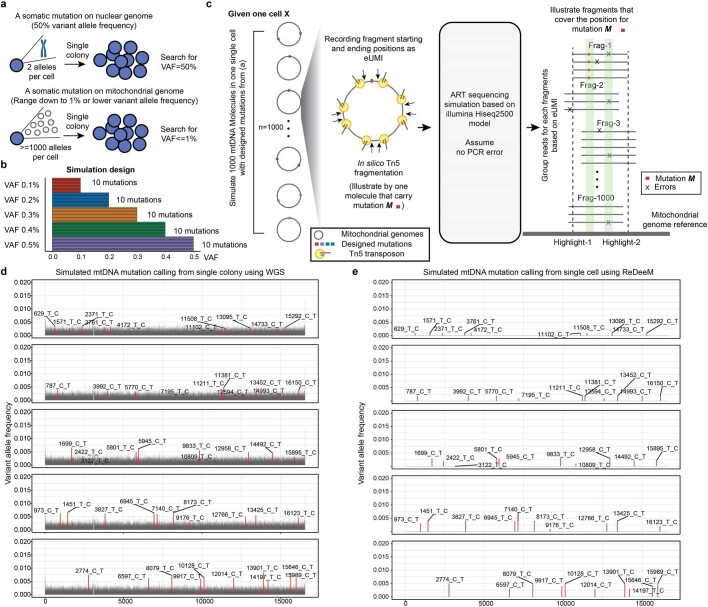
Extended Data Fig. 2. eUMI-based error correction via ReDeeM versus conventional mutation detection.
a, The challenge of mtDNA mutation calling using conventional WGS in single colony. b-e, Simulation analysis of mtDNA mutation calling using WGS vs ReDeeM. b, The design of the mtDNA mutations with low heteroplasmy (0.1% ~ 0.5%) for simulation analysis. 10 mutations are randomly picked for each variant allele frequency (VAF). c, Illustration of the simulation analysis process. One single cell with 1000 mtDNA copies is simulated, with the designed mutations from panel a. Next, in silico Tn5 fragmentation followed by artificial sequencing is simulated. The resulting simulated data is analyzed by ReDeeM or conventional mutation calling pipeline. The highlight-1 (Real mutation M) and highligh- 2 (Error) have the same total frequency which can only be distinguished by ReDeeM. d-e, Mutation calling results using conventional WGS in d and the eUMI-based ReDeeM pipeline in e. Also see Supplementary Notes.