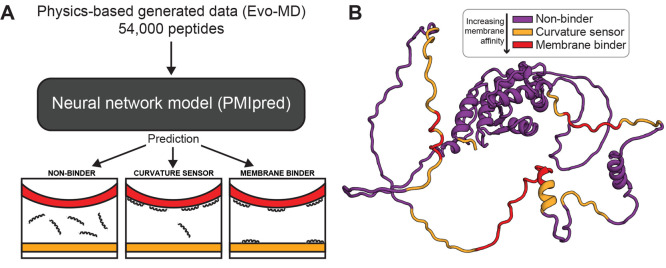
Figure 5.
Overview of PMIpred: on-the-fly prediction of peptide–membrane interaction using physics-based inverse design. Adapted with permission from ref (19). Copyright 2023 The Authors, preprinted by Cold Spring Harbor Laboratory. (A) Data produced using physics-based inverse design approaches such as Evo-MD can be used to train deep learning models, allowing for quick but accurate evaluation of entire proteins while avoiding the overhead of MD simulations. PMIpred incorporates a transformer model trained on Evo-MD generated data for optimizing curvature sensing, allowing for the classification of peptides and regions of proteins as either nonbinding, curvature sensing, or membrane binding. (B) Example output of PMIpred showing the protein structure of ArfGAP1. Regions of interest are labeled according to the model, indicating regions likely to exhibit curvature sensing, membrane binding, or nonbinding behavior.