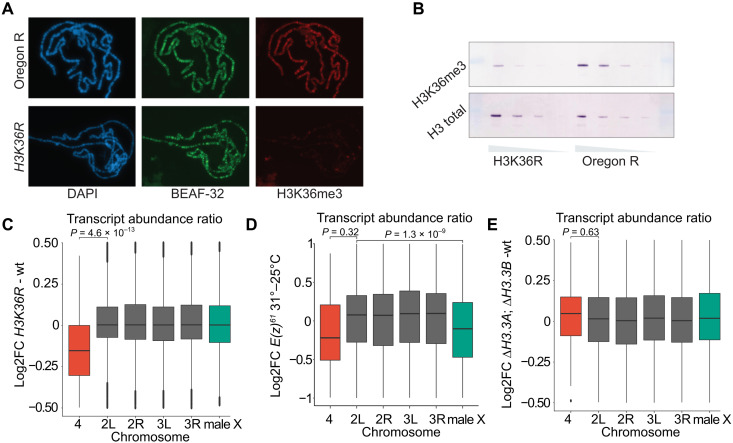
Fig. 3. Set2 contributes to increased transcriptional output from the male X chromosome independently of H3K36 methylation.
(A) Polytene chromosome staining of H3K36me3 in wild type (top row) and ΔHisC; 12xH3K36R histone replacement larvae (bottom row). Staining of boundary element-associated factor of 32kD (BEAF-32) serves as an internal control. (B) Western blot analyses of twofold dilutions of total protein lysates from brains and imaginal discs of third instar larvae show substantial decrease of H3K36me3 (top row) in ΔHisC; 12xH3K36R compared to wild type. Western blot with antibodies against total histone H3 serves as loading control. (C) Box plots of log2 fold change of transcript abundance per chromosome for ΔHisC; 12xH3K36R versus ΔHisC; 12xH3K36K (wt). Note that chromosome 4 shows a significant reduction in transcript abundance compared to the other chromosome arms. Here and below, unpaired two-sample Wilcoxon tests were used to determine the likelihood that observed differences are explained by chance. (D) Box plots of log2 fold change of transcript abundance per chromosome for the E(z)61 cell line at 31°C [strongly reduced E(z) function] versus 25°C. At restrictive temperature, the abundance of transcripts from X chromosome is significantly reduced compared to the major autosome arms. (E) Box plots of log2 fold change of transcript abundance per chromosome for ΔH3.3B; ΔH3.3A versus wild-type flies indicate no significant differences.