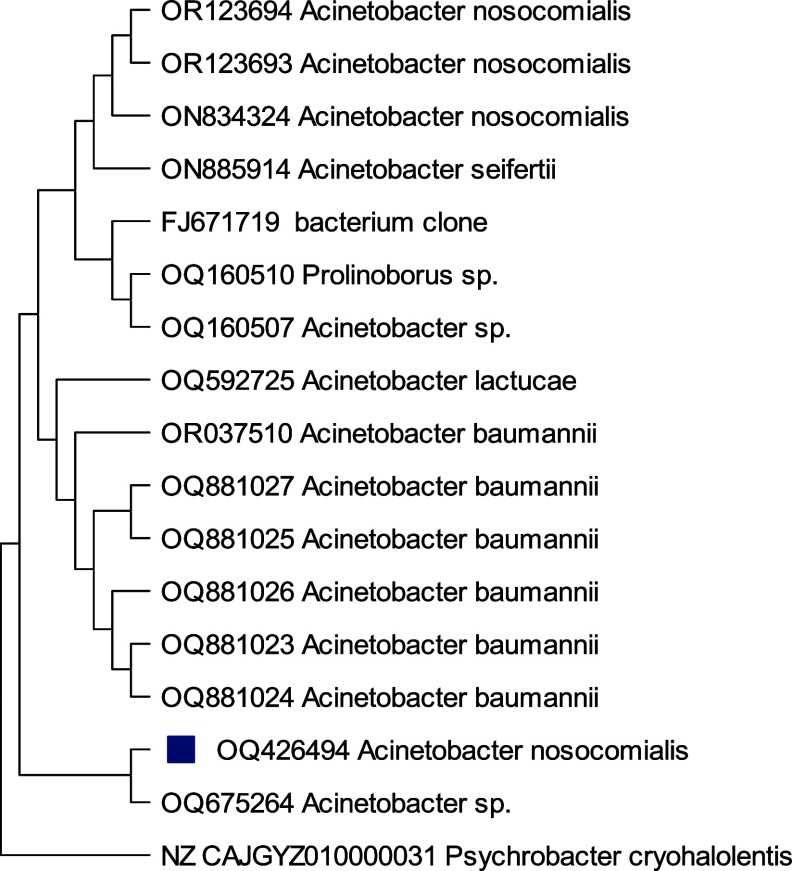
Figure 9.
Phylogenetic tree of Acinetobacter nosocomialis H22 by the neighbor-joining method with the bootstrap test showing replicate percentage and associated taxa. The tree with the highest log likelihood (−3730.65) is shown. Maximum Composite Likelihood tool computed evolutionary distances with 31 nucleotide sequence analysis by deletion of ambiguous pair wise positions. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 0.5376)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 37.37% sites). Final data set has 1504 positions.