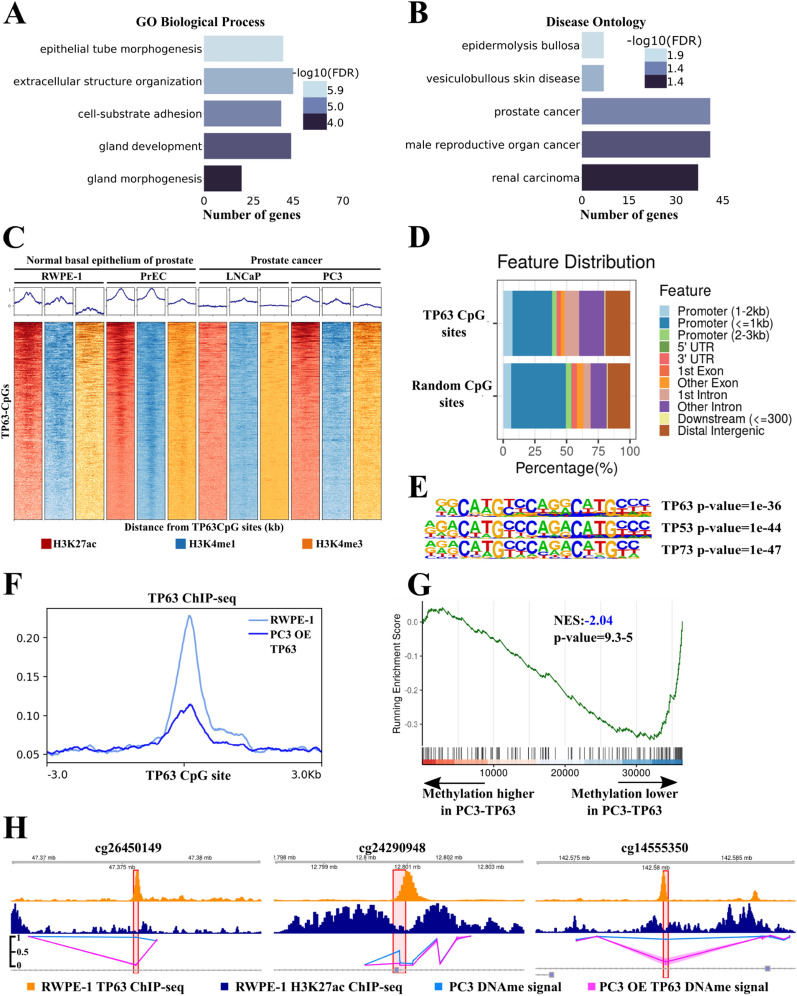
Fig. 3.
CpG sites associated with TP63 cluster lie in TP63-dependent enhancers and super-enhancers. A, B Gene Ontology (GO) analysis of genes located close to TP63 CpG sites. GO terms related to biological process (A) and disease ontology (B) are shown. FDR is indicated with a color scale. C ChIP-seq density plots of H3K27ac, H3K4me1, and H3K4me3 enrichments at TP63 CpG sites in the RWPE-1, PrEC, LNCaP, and PC3 cell lines. Each column represents a 6-kb interval centered on a CpG site. D Bar plot of genomic distribution of the TP63 CpG sites (top) and random CpG sites (bottom). E Representation of p53 protein family motifs enriched at TP63 CpG sites using HOMER. F Mean TP63 ChIP-seq signal at TP63 CpG sites in the RWPE-1 cell line and PC3 with overexpression (OE) of TP63. G GSEA-like plot for methylation data evaluating the TP63 CpG sites signature upon TP63 overexpression in PC3. H ChIP-seq profiles of H3K27ac (top track) and TP63 (middle track) in RWPE-1 cell line and DNA methylation profile in PC3 cell line (bottom track) following overexpression (OE) of TP63 or GFP as a control at three CpG sites