Figure 5. Linking prioritized AD variants to phenotypes by allelic analyses and cellular functional assays.
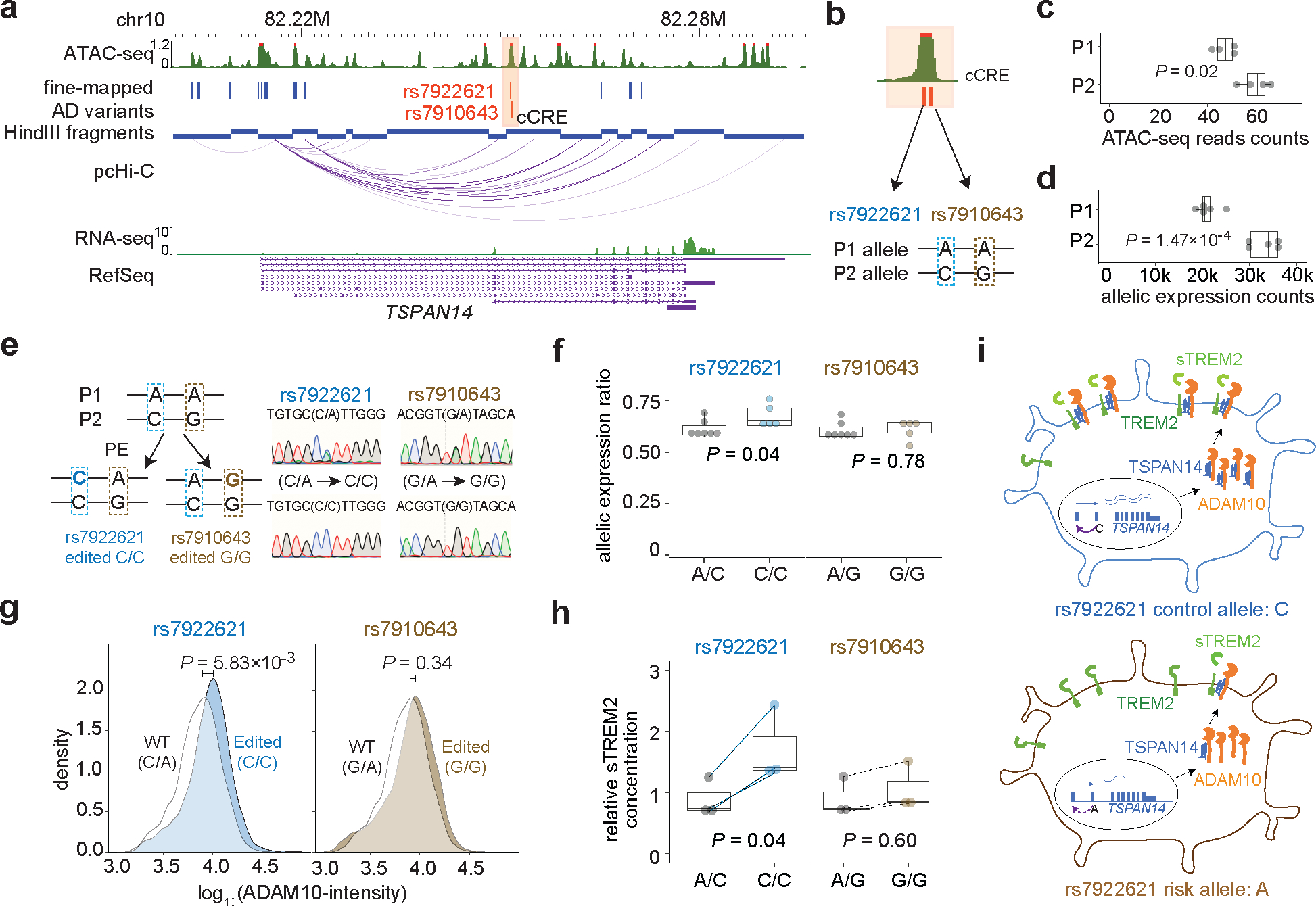
a, Two prioritized AD variants located in a cCRE (highlighted in orange) physically interacting with TSPAN14 promoter. b, hESC H1 genome is heterozygous for the two AD variants. The P1 allele has the risk alleles: rs7922621 (A) and rs7910643 (A), while the P2 allele has the non-risk alleles: rs7922621 (C) and rs7910643 (G). c, Allelic analysis of ATAC-seq data in H1 derived microglia reveals decreased chromatin accessibility of the P1 allele compared to the P2 allele (two-sided binomial test, n = 4). d, Allelic analysis using haplotype-resolved SNPs in the TSPAN14 gene body shows reduced TSPAN14 expression from the P1 allele compared to the P2 allele in microglia (two-sided binomial test, n = 5). e, Illustration of the prime editing strategy to convert rs7922621 (A) and rs7910643 (A) on the P1 allele to rs7922621 (C) and rs7910643 (G), respectively. Representative results from sanger sequencing display wildtype clones and KI clones. f, Allelic imbalance of TSPAN14 gene expression in the wild type clones (A/C) is partially reduced by prime-editing of rs7922621 (A/C to C/C) but not rs7910643. P values calculated using two-sided two-sample t-test (n = 5). g, Microglia with rs7922621 (C/C) genotype have elevated cell surface ADAM10 than wildtype microglia by immunostaining and FACS analysis. P values calculated using one-sided (edited > WT) two-sample Wilcoxon test (n = 5). h, rs7922621 (C/C) prime-edited microglia, but not rs7910643 (G/G) edited microglia, shed significantly more sTREM2 than wildtype microglia. P values calculated using two-sided paired (dash line) t-test between wildtype and prime-edited microglia from the same differentiation batch (n = 3). Boxplot indicates the median and interquartile range and whiskers mark the 5th and 95th percentiles for (c), (d), (f), (h). I, Proposed model of linking AD risk variant rs7922621 to function. rs7922621 risk allele A disrupts cis-regulatory function and down-regulate TSPAN14 expression, which leads to impaired ADAM10 trafficking and maturation to cell surface. The reduced ADAM10 level at the cell surface reduces TREM2 cleavage and sTREM2 shedding.
