Extended Data Fig. 3. Enrichment analysis of IFNβ responsive genes compared to disease-associated microglia (DAM) feature genes.
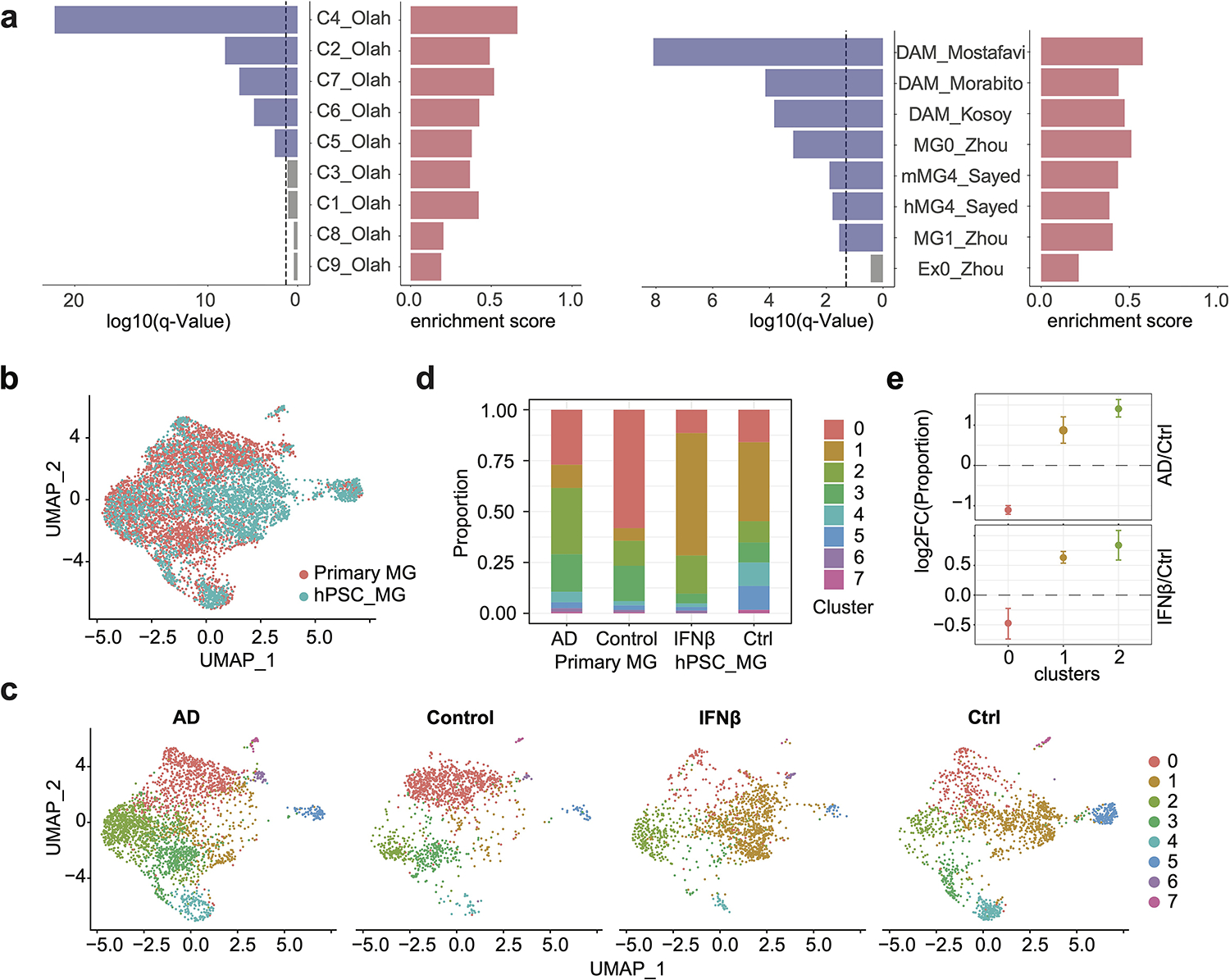
a, Barplots show the q values and enrichment scores of GSEA results for IFNβ responsive genes, in comparison with published datasets17,18,23–26 (dash line, q = 0.05). IFNβ responsive genes are highly enriched in multiple clusters of DAM by Olah et al.18, including the C4 cluster, representing cells with activated IFN signaling, the C7 cluster, representing cells expressing DAM genes, as well as C5 and C6, representing cells expressing genes related to anti-inflammatory responses. The C2 cluster, representing homeostatic microglia which are more likely derived from the temporal neocortex of younger temporal lobe epilepsy patients compared to the other homeostatic population, is also enriched for IFNβ responsive genes. 4 clusters are not enriched for IFN responsive genes, including C1, a homeostatic population shared by all brain regions in all donors, C3, cells with enriched expression of cellular stress genes, C8, cells enriched for respiratory electron transport, and C9 enriched with genes of cell cycle. In addition, IFNβ responsive genes are enriched in microglia samples associated with AD from 5 additional studies, including microglia samples in the human-MG4 cluster and the mouse-MG4 cluster, which are most enriched with DAM genes among all clusters from Sayed et al.17, the MG0 cluster (highly represented in AD microglia) compared to the MG1 cluster (control microglia) from Zhou et al.23, and AD DAM DEGs from Mostafavi, et al.24, Kosoy et al.26, and Morabito et al.25. b, UMAP plot visualizing integration of 3,038 WTC11-microglia scRNA-seq with 4,126 primary microglia snRNA-seq from Morabito et al. Cells are colored by sample origins. c, UMAP plot visualizing joint clustering splitted by donor condition (AD/control) or treatment (IFNβ stimulation/control). d, Cell proportions of each cluster splitted by donor condition or treatment. e, Cell proportion fold change in AD vs control or IFNβ stimulation vs control for 3 major clusters using monte-carlo/permutation test. Data are shown as mean ± s.d. (n_permutations = 1000).
