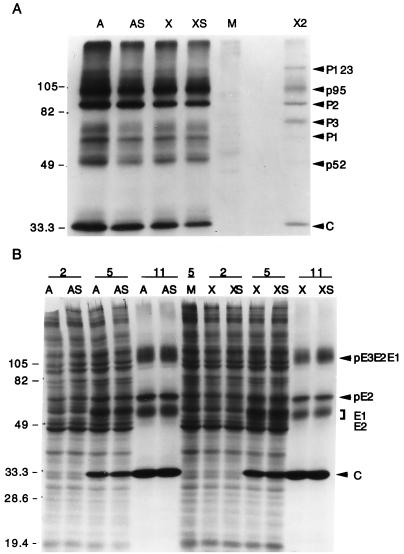
FIG. 5.
CrmA does not alter production of nonstructural and structural Sindbis virus proteins. (A) Sindbis virus nonstructural proteins (P123, P1, P2, and P3) were immunoprecipitated from equal volumes of labeled lysates, prepared 11 h after infection, with recombinant viruses encoding crmA (A), crmA-stop (AS), bcl-xL (X), and bcl-xL-stop (XS) or from mock-infected cells (M) with rabbit antiserum raised against nsP2. The nonstructural proteins were identified by their molecular masses (the positions of molecular size markers [in kilodaltons] are shown on the left), by comparison to published protein patterns (15), and by comparison to nonstructural proteins immunoprecipitated with a mixture of antibodies to nsP2 and nsP3 (X2) provided by M. Gorrell and D. Griffin. Proteins were resolved by SDS–10% PAGE and processed with salicylic acid prior to autoradiography. The results are representative of three independent experiments. (B) Sindbis virus structural proteins were analyzed by SDS–15% PAGE and autoradiography of labeled whole-cell lysates harvested at the indicated times (in hours) after infection of BHK cells. These results are representative of six independent time course experiments. The arrows indicate the precursor viral glycoproteins (pE3E2E1 and pE2), the mature glycoproteins (E1 and E2), and the capsid protein (C). Molecular mass standards (in kilodaltons) are indicated.