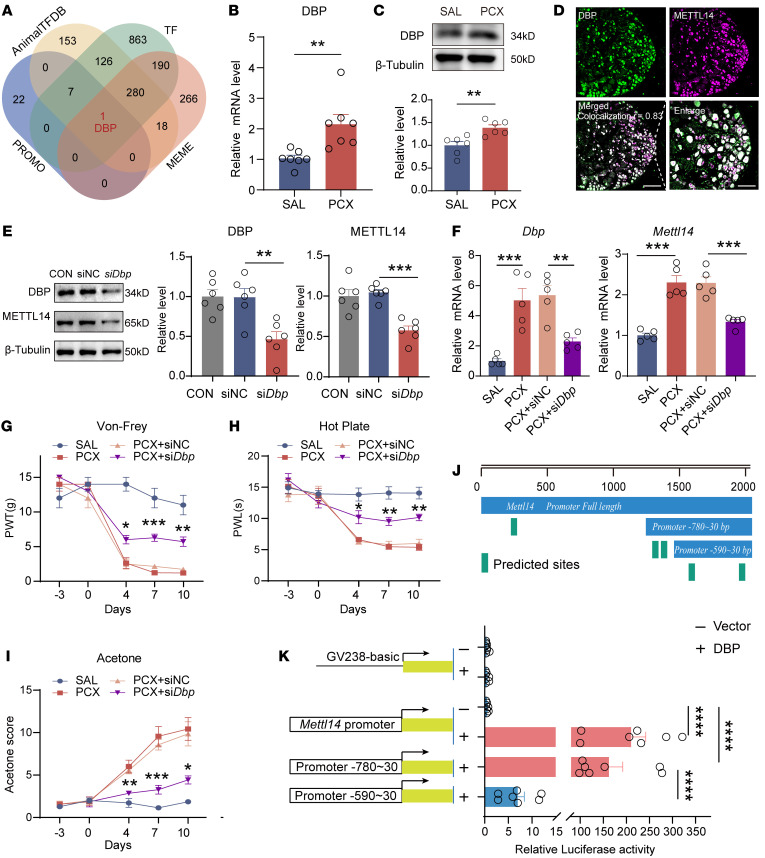
Figure 10. The transcription factor DBP mediates METTL14 upregulation caused by paclitaxel treatment.
(A) Venn diagram shows the predictive transcription factors (TFs) by taking intersections with the TFs of the rat species and predictive results from the PROMO, AnimalTFDB, and MEME (FIMO function) databases. (B and C) Dbp mRNA (B) and protein (C) expression in the bilateral L4–L6 DRGs in saline- or PCX-pretreated rats (at least 6 rats per group, Student’s t test). (D) Colocalization analysis of DBP and METTL14 based on double-labeled immunofluorescent images. Scale bars: 100 μm, left; 50 μm, enlarged image. (E) DBP and METTL14 protein expression in PC12 cells transfected with siDbp compared with the control (n = 6 per group, 1-way ANOVA followed by Tukey’s post hoc test). (F) Dbp and Grin2a mRNA expression in the rat bilateral L4–L6 DRGs in saline- or PCX-treated rats with siRNA-Mettl14 or scrambled siRNA injection (n = 5 rats per group, 1-way ANOVA followed by Tukey’s post hoc test). (G–I) Mechanical allodynia, thermal hyperalgesia, and cold allodynia of saline- or PCX-treated rats after siRNA-Dbp or scrambled siRNA treatment (at least 7 rats per group, 2-way ANOVA followed by Tukey’s post hoc test). (J) Top: Schematic representation of the full-length Mettl14 promoter, truncation promoters (–780 bp to ~30 bp, –590 bp to ~30 bp), and predicted binding sites from the JASPAR database. Bottom: Vectors containing full-length Mettl14 promoter and Mettl14 truncation promoters (–780 bp to ~30 bp) show higher luciferase activity (at least n = 7 per group, 1-way ANOVA followed by Tukey’s post hoc test). Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.