Extended Data Fig. 6. Editing specificity of monomeric mitoABEs.
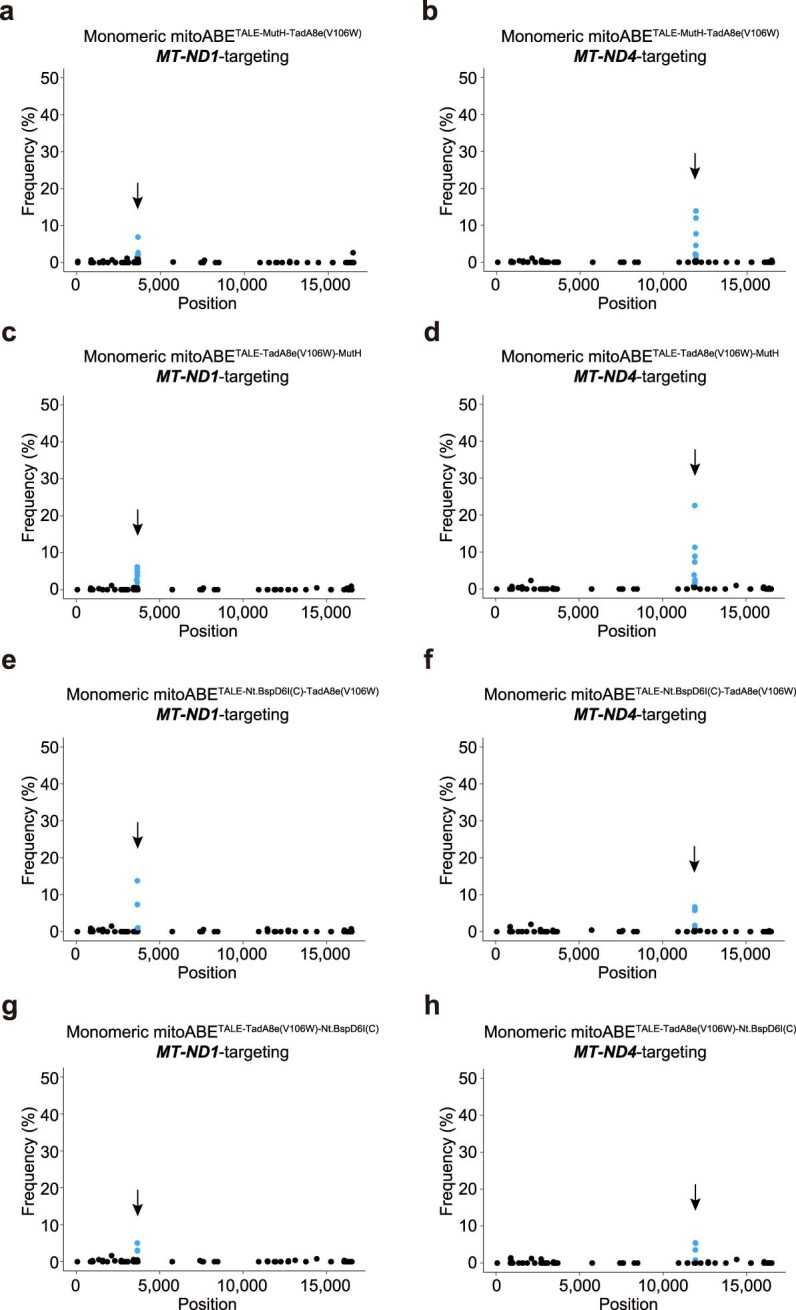
a-h, The average frequency and mitochondrial genome position of each unique single nucleotide variant (SNV) are shown for MT-ND1-targeting monomeric mitoABEMutH (TALE-MutH-TadA8e(V106W)) (a), MT-ND4-targeting monomeric mitoABEMutH (TALE-MutH-TadA8e(V106W)) (b), MT-ND1-targeting monomeric mitoABEMutH (TALE-TadA8e(V106W)-MutH) (c), MT-ND4-targeting monomeric mitoABEMutH (TALE-TadA8e(V106W)-MutH) (d), MT-ND1-targeting monomeric mitoABENt.BspD6I(C) (TALE-Nt.BspD6I(C)-TadA8e(V106W)) (e), MT-ND4-targeting monomeric mitoABENt.BspD6I(C) (TALE-Nt.BspD6I(C)-TadA8e(V106W)) (f), MT-ND1-targeting monomeric mitoABENt.BspD6I(C) (TALE-TadA8e(V106W)-Nt.BspD6I(C)) (g), MT-ND4-targeting monomeric mitoABENt.BspD6I(C) (TALE-TadA8e(V106W)-Nt.BspD6I(C)) (h). a-h, all data presented as mean values of n = 3 independent biological replicates. The arrow points to the targeted editing site and the blue dots represent the editing efficiency of adenines in the editing window.
