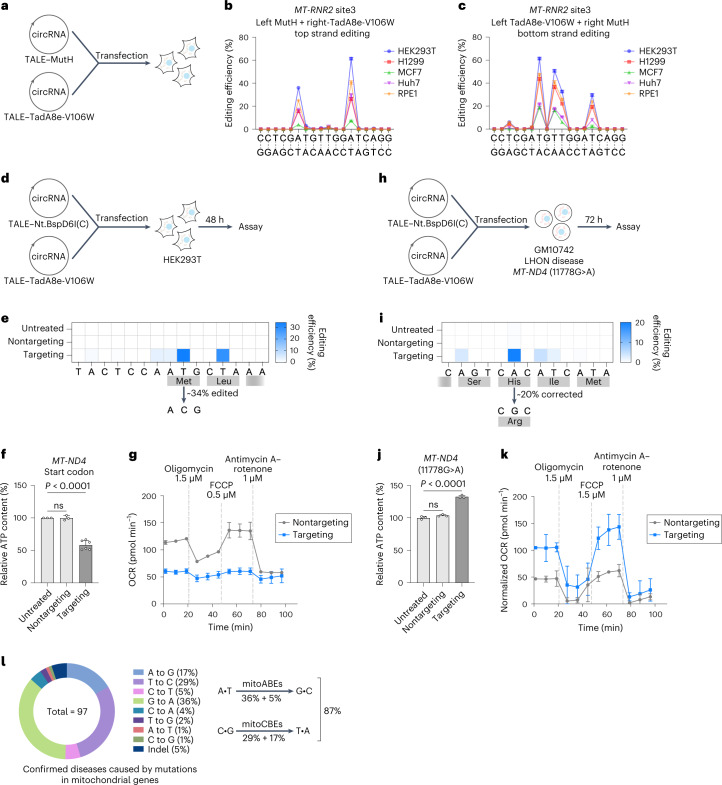
Fig. 6. circRNA-encoded mitoABEs successfully created disease models and corrected mutation in cells derived from individuals with LHON.
a, Overview of circRNA-encoded mitoABEMutH-transfected cells. b,c, circRNAs of two mitoABEMutH orientations were transfected into different cell lines to achieve strand-biased editing, left TALE–MutH with right TALE–TadA8e-V106W (b), left TALE–TadA8e-V106W with right TALE–MutH (c). Genomic DNA was collected 2 days posttransfection. d, Overview of circRNA-encoded mitoABENt.BspD6I(C)-transfected HEK293T cells and genomic DNA collected 2 days posttransfection. e, The editing efficiencies of circRNA-encoded mitoABENt.BspD6I(C) targeted the start codon of MT-ND4. f, The ATP levels of cells transfected with circRNA-encoded mitoABENt.BspD6I(C) targeted the start codon of MT-ND4. Student’s t test, P = 2.71 × 10−5. g, Oxygen consumption rate (OCR) in HEK293T cells treated with circRNA-encoded mitoABENt.BspD6I(C) targeted the start codon of MT-ND4 for 2 days. h, Overview of circRNA-encoded mitoABENt.BspD6I(C)-transfected LHON disease cells GM10742, with genomic DNA was collected 3 days posttransfection. i, The editing efficiency of mitoABENt.BspD6I(C) corrected the 11778G>A mutation of LHON disease cell GM10742. j, The ATP levels of cells transfected with circRNA-encoded mitoABENt.BspD6I(C) targeting the 11778G>A mutation of LHON disease cell GM10742. Student’s t test, P = 6.98 × 10−5. k, OCR of the LHON disease cell line GM10742 treated with circRNA-encoded mitoABENt.BspD6I(C) targeting the 11778G>A mutation for 2 days. l, Types of mitochondrial diseases (MITOMAP) and the proportion of diseases that can theoretically be treated by mitoBEs. For b, c, f, g, j and k, the data are presented as mean values ±s.d. of n ≥ 3 independent biological replicates. For e and i, the mean values from n = 3 biologically independent replicates are shown. For g,k, FCCP represents carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone.