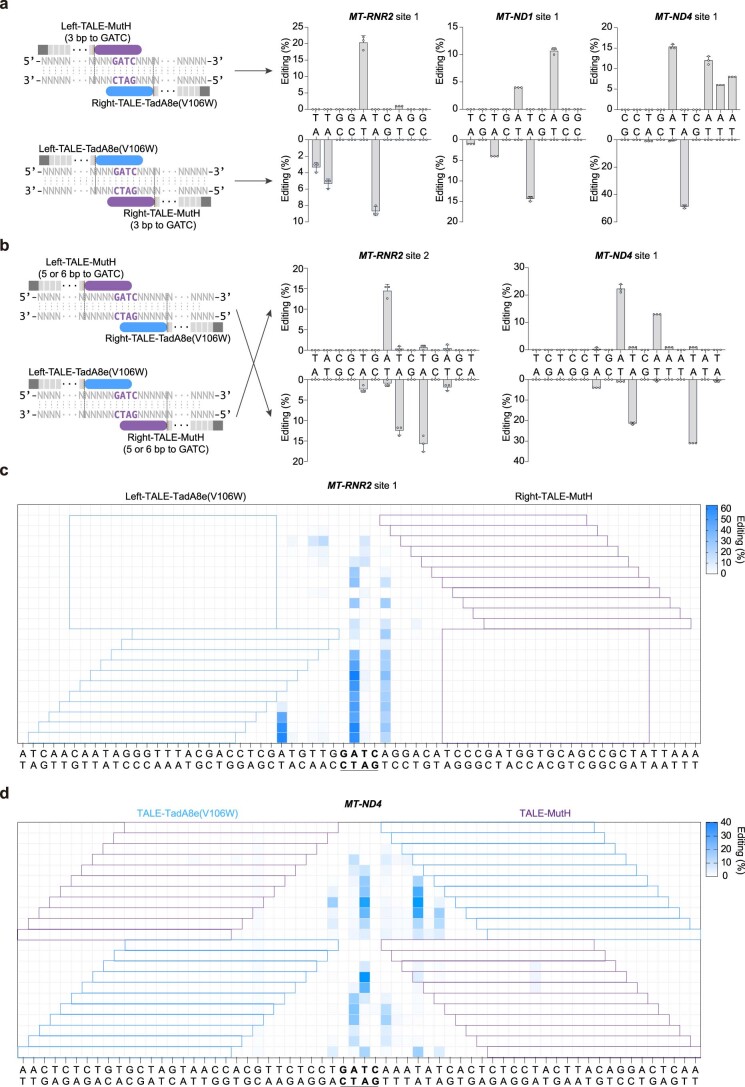
Extended Data Fig. 2. Strand-biased editing of mitochondrial DNA using mitoABEMutH.
a, Different orientations of mitoABEMutH when the 3 bp distance from 5’-GATC-3’ and the adenines on different strands are edited at MT-RNR2 site 1, MT-ND1 site 1 and MT-ND4 site 1. Top, Left-TALE-MutH and Right-TALE- TadA8e(V106W). Bottom, Left-TALE-TadA8e(V106W) and Right-TALE- MutH. b, Different orientations of mitoABEMutH when the 5 or 6 bp distance from 5’-GATC-3’ and the adenines on different strands are edited at MT-RNR2 site 2 and MT-ND4 site 1. Top, Left-TALE-TadA8e(V106W) and Right-TALE-MutH. Bottom, Left-TALE-MutH and Right-TALE-TadA8e(V106W). For a and b, data are presented as mean values ± s.d. of n = 3 independent biological replicates. In a and b, the blue rounded rectangle represents TadA8e(V106W), the purple rounded rectangle represents nickase, and the gray rectangle represents TALE. c and d, Editing efficiency of mitoABEMutH with MutH and TadA8e(V106W) at different distances from 5’-GATC-3’ at MT-RNR2 site 1 (c) and MT-ND4 (d). For c and d, the mean values from n = 3 biologically independent replicates are shown.