Summary
Here, we present a protocol for the identification of differentially expressed genes through RNA sequencing analysis. Starting with FASTQ files from public datasets, this protocol leverages RumBall within a self-contained Docker system. We describe the steps for software setup, obtaining data, read mapping, sample normalization, statistical modeling, and gene ontology enrichment. We then detail procedures for interpreting results with plots and tables. RumBall internally utilizes popular tools, ensuring a comprehensive understanding of the analysis process.
Subject areas: Bioinformatics, Sequence analysis, RNA-seq
Graphical abstract

Highlights
-
•
The protocol necessitates the setup of Docker, RumBall, and a command line interface
-
•
External datasets for reference and annotation can be easily obtained using built-in scripts
-
•
Identification of DEGs achievable in a few steps
-
•
Steps outlined for simplifying scripting and tool management within a Docker container
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present a protocol for the identification of differentially expressed genes through RNA sequencing analysis. Starting with FASTQ files from public datasets, this protocol leverages RumBall within a self-contained Docker system. We describe the steps for software setup, obtaining data, read mapping, sample normalization, statistical modeling, and gene ontology enrichment. We then detail procedures for interpreting results with plots and tables. RumBall internally utilizes popular tools, ensuring a comprehensive understanding of the analysis process.
Before you begin
This protocol describes the basic usage of the RumBall for bulk RNA-seq analysis.1 It serves as an extensive guide for users aiming to conduct RNA-seq analysis in a manner that is both reproducible and straightforward. The primary objective is to demystify the often-complex steps involved in analyzing RNA-seq data, thereby making the process accessible to researchers from various backgrounds.
Previous efforts have materialized useful computational platforms designed to streamline the intricacies of RNA-seq analysis for researchers, using systems such as Nextflow,2 Snakemake,3 and web server systems.4,5,6,7,8 However, the complexity of analyzing multiple RNA-seq datasets coupled with challenging software setups discourages non-experts. Furthermore, the server dependencies of web tools and their input limitations hinder full-scale analysis starting from FASTQ files.
To address these problems, we introduce RumBall: a user-friendly, scalable, and reproducible platform that packages state-of-the-art tools for comprehensive bulk RNA-seq analysis into a virtual kit using container technology. This enables automated, version-controlled analyses that can be executed on any system from FASTQ files to functional analysis.
It is important to note, however, that RumBall is neither faster nor more advanced than existing RNA-seq platforms. Its design prioritizes functionality, portability, and reproducibility, requiring minimal setup and commands. For users who require massive scaling for extensive RNA-seq datasets – numbering in the thousands – alternative solutions may be more suitable. These alternatives, while offering vast scalability, come with a steeper learning curve and more complex dependency requirements.
To run this tutorial, only a command-line interface, Docker, and RumBall are required. For customization, users need a basic knowledge of the Linux/Bash command line. Even non-specialists might find the explanations comprehensible and learn how to adapt the pipeline for original data analysis on their desired datasets. The simplicity of this approach is achieved by having all tools pre-configured and ready to run within the container.
Install Docker
Timing: 5 min
Docker is a platform that encapsulates the necessary computational tools, packages, and libraries into containers.9 This platform enables users to share and run identical computing environments across various machines, including high-performance servers and laptop PCs.
Below, we provide the commands for installing Docker on Ubuntu Linux as an example. For other operating system, follow the installation instructions on the Docker website (https://docs.docker.com/engine/install/).
Note: This step can be skipped if Docker is already available on your system.
Note: The superuser (sudo) permission is required for this step. If you do not have the permission, it is advisable to contact the administrator of your system.
-
1.Refresh the list of available software and their details.
-
a.Install the necessary tools so that our software management system can securely fetch software from online storage locations:
-
a.
>sudo apt-get update
>sudo apt-get install ca-certificates curl gnupg
-
2.
Add Docker’s official GPG key.
>sudo install -m 0755 -d /etc/apt/keyrings
>curl -fsSLhttps://download.docker.com/linux/ubuntu/gpg∖
| sudo gpg --dearmor -o /etc/apt/keyrings/docker.gpg
>sudo chmod a+r /etc/apt/keyrings/docker.gpg
Note: GPG stands for GNU Privacy Guard, which is a tool for secure communication and data storage. A GPG key is used to encrypt and sign digital content. In the context of software and package repositories, it ensures that the software you're downloading is authentic and hasn't been tampered with by any malicious third parties.
-
3.
Set up the repository (where and how to find the software library) and update the apt-package index:
>echo
"deb [arch="$(dpkg --print-architecture)" signed-by=/etc/apt/keyrings/docker.gpg]https://download.docker.com/linux/ubuntu∖
"$(. /etc/os-release && echo "$VERSION_CODENAME")" stable" ∖
| sudo tee /etc/apt/sources.list.d/docker.list > /dev/null
>sudo apt-get update
-
4.
Install Docker Engine:
>sudo apt-get install docker-ce docker-ce-cli containerd.io docker-buildx-plugin docker-compose-plugin
-
5.
Initialize Docker and verify that Docker is successfully installed.
>sudo service docker start
>sudo docker run hello-world
-
6.Create the docker group.Note: Docker commands typically require sudo permission. To avoid prefixing every Docker command with ‘sudo,’ you can create a new group that the system trusts and then add users to that group.
-
a.Create the group:>sudo groupadd docker
-
b.Add the user to the group:>sudo usermod -aG docker $USER
-
c.Restart the computer or activate the changes through the command:>newgrp docker
-
d.Test the Docker command without sudo permission:>docker --version
-
a.
Obtaining RumBall
Timing: 12 min
-
7.
Download RumBall docker image from the DockerHub repository (https://hub.docker.com/r/rnakato/rumball):
> docker pull rnakato/rumball
-
8.
To confirm that the image was successfully downloaded, run one of the scripts contained in RumBall:
> docker run --rm -it rnakato/rumball star.sh
Note: RumBall is continuously updated to incorporate the latest advances in transcriptome analysis. It is therefore recommended to check our recent updates at the web manual page https://rumball.readthedocs.io/en/latest/index.html.
-
9.
To download a specific version of RumBall, enter the following command replacing the <version> field to the desired version:
> docker pull rnakato/rumball:<version>
-
10.
To execute commands in specific version:
> docker run --rm -it rnakato/rumball:<version> ls
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| FASTQ files | Zuin et al.10 | GEO: GSE44267 (GSM1081538, GSM1081539, GSM1081540, GSM1081541) |
| Software and algorithms | ||
| Docker | Docker Inc. | https://www.docker.com/ |
| RumBall | DockerHub | https://hub.docker.com/r/rnakato/rumball |
| Other | ||
| Workstation | HP | HP ML350p |
Materials and equipment
Computer hardware
For this tutorial, we tested it on Ubuntu Server 22.04 using a workstation with 32 Intel Xeon CPUs and 64 GB RAM, with an internet connection. The whole analysis including all produced files will occupy approximately 40 GB of the computer’s hard disk. We recommend ensuring approximately 64 GB of storage is available before starting this protocol.
Software versions
As stated in the before you begin section, you will need a command-line interface compatible to your operating system, Docker, and RumBall. All necessary tools are already preconfigured and housed within RumBall.
The version of RumBall used in this protocol (v.0.5.0) has the following tools preinstalled.
Mapping tools
Mapping tools for RNA-seq
RNA-seq tools
-
•
RSEM18 v1.3.3.
-
•
edgeR19 v3.38.1.
-
•
DESeq220 v1.36.0.
-
•
stringtie21 v2.2.1.
-
•
Ballgown22 v2.18.0.
-
•
sleuth23 v0.30.0.
Gene ontology (GO) analysis tools
Utility tools
Pause point: In the course of this protocol, you will encounter several pause points. These checkpoints involve saving files to your hard disk. Detailed information about these outputs is provided in the expected outcomes section.
Step-by-step method details
This tutorial covers four main processes: (1) preparing the datasets or downloading from SRA, (2) checking the strandedness of RNA-seq data, (3) mapping reads and estimate the gene expression level for each sample, and (4) analyzing differential gene expression. We explain each step in detail, along with the corresponding codes.
Preparing datasets and reference genome
Timing: 1–8 h
We perform RNA-seq analysis on four RNA-seq samples (GEO: GSE44267) from immortalized human embryonic kidney cells (HEK293), as detailed in Zuin et al.10
Note: We break down the explanations line by line, so that users can understand and learn how to modify the script on their own when needed. Repetitive parts will be skipped in subsequent explanations.
-
1.
Create a project directory to store all the analysis files with ‘mkdir’ and then navigate into it with the cd command:
>mkdir RumBall_tutorial
>cd RumBall_tutorial
Pause point: There are two pathways to proceed from this point. If users possess their own datasets in FASTQ format, they should proceed to step 2. Alternatively, if users wish to utilize the same datasets in this tutorial, they should continue with step 3.
-
2.Prepare own FASTQ and metadata files:
-
a.Create fastq directory:>mkdir fastq
-
b.Navigate to the directory containing the FASTQ files:>cd /path/to/fastq-files
-
c.Next, instead of copying or moving your original datasets, we recommend creating symbolic links (symlinks) to the fastq directory. Symlinks points to another file or directory on the file system, acting like a shortcut without duplicating the actual data.>for file in ∗.fastq; do> ln -s "$(pwd)/$file" /path/to/project/fastq/$file>doneNote: ‘for file in ∗.fastq; do’: This initiates a for loop, which will iterate over every file in the current directory that ends with the ‘.fastq’ extension.Note: Include ‘.gz’ if it is applicable to your dataset. The variable file will hold the name of each FASTQ file one by one as the loop progresses.Note: ‘ln -s "$(pwd)/$file" /path/to/destination/$file’: This command creates a symlink for each FASTQ file. ‘ln -s’ is the command to create a symbolic link. ‘"$(pwd)/$file"’ is the source file for the symlink. ‘$(pwd)’ generates the full path of the current directory, and ‘$file’ is the name of the current FASTQ file being processed by the loop. Together, they form the full path to the source file. ‘/path/to/destination/$file’ specifies where the symlink will be created. This should be replaced with the actual path where you want the symlinks to be located. ‘$file’ at the end ensures that each symlink has the same name as the original FASTQ file. ‘done’: This marks the end of the for loop.
-
d.Return to the project directory and list (using the command ls) the files to confirm the contents:>cd /path/to/project/fastq/>ls
 CRITICAL: We assume the naming convention ‘<ID>_1.fastq.gz’ and ‘<ID>_2.fastq.gz’ for Read1 and Read2, respectively. If your files follow a different pattern, please rename them accordingly using the command ‘mv’ with first the original name and then the desired name:>mv sample1_control_R1.fq.gz sample1_1.fastq.gz
CRITICAL: We assume the naming convention ‘<ID>_1.fastq.gz’ and ‘<ID>_2.fastq.gz’ for Read1 and Read2, respectively. If your files follow a different pattern, please rename them accordingly using the command ‘mv’ with first the original name and then the desired name:>mv sample1_control_R1.fq.gz sample1_1.fastq.gz -
e.For the metadata file, we recommend users following our example below:>cat >sample_info.csv <<EOF>sample1,biosample_Control_rep1,control>sample2,biosample_Control_rep2,control>sample3,biosample_Exp_rep1,KO>sample4,biosample_Exp_rep2,KO>EOFNote: Here we create a file named ‘sample_info.csv’ where each line represents a sample. ‘cat’ is a command for concatenation, and it can also read and write files. The ‘>’ operator redirects the output to the ‘sample_info.csv’ file. ‘EOF’ stands for ‘End Of File’ and it is used a delimiter in shell scripts to denote the beginning and end of the document we are redirecting line by line to the ‘sample_info.csv’ file. Please add a simple comma (‘,’) without any space in between fields. It is also recommended to use underscore (‘_’) for separations in the sample name.Note: For sample names, we recommend an informative and clear naming convention applied to all samples. Here we use the format: ‘<biological sample>_<experiment>_<replicate>’. However, the necessary content should be discussed and defined in your research group. For example, in case samples are collected from different sequencing technology, or have varying read length, and so on.
-
a.
-
3.In case users do not possess own datasets, download FASTQ files and prepare the metadata file outlined below:
-
a.Download from public database Sequence Read Archive (SRA)26:>rumball="docker run --rm -it --user $(id -u):$(id -g) -v $(pwd):/work rnakato/rumball">mkdir -p fastq>$rumball fasterq-dump SRR710092 -O /work/fastq -e 16 -p>$rumball fasterq-dump SRR710093 -O /work/fastq -e 16 -p>$rumball fasterq-dump SRR710094 -O /work/fastq -e 16 -p>$rumball fasterq-dump SRR710095 -O /work/fastq -e 16 -p
 CRITICAL: It will take several hours to complete. Do not close the terminal window until it finishes.Note: The exact duration of this step primarily depends on the user’s computational processing power and internet connection speed.Note: ‘rumball = "docker run --rm -it --user $(id -u):$(id -g) -v $(pwd):/work rnakato/rumball"’ sets up an alias called ‘rumball’ that will run a Docker container based on the image ‘rnakato/rumball.’ The options ‘--rm’ and ‘-it’ ensure that the container is removed after exiting and that it runs interactively, respectively. The option ‘--user $(id -u):$(id -g)’ specifies the user and groups IDs to use inside the container, ensuring that the container runs with the same user/group permissions as the host user. The option ‘-v $(pwd):/work’ mounts the current working directory on the host machine ‘($(pwd))’ to the ‘/work’ directory inside the container. This allows data to be shared between the host and the container. The command ‘mkdir -p fastq’ creates a new directory named ‘fastq’ if it does not already exist (with ‘-p’). The command ‘$rumball fasterq-dump SRR710092 -O fastq -v’ executes the ‘fasterq-dump’ command from the SRA-Toolkit inside the RumBall Docker container to download the SRA file with ID SRR710092 into the directory fastq (with ‘-O’). Fasterq-dump assumes each SRA contain paired-end reads (‘--split-files’) by default. To improve the speed of file conversion, the parameter ‘-e’ followed by number of threads can allocate more CPU cores to the task and ‘-p’ (progress) provides more detailed information upon completion. You can assess the number of cores in your system by typing ‘nproc --all’ on your Linux terminal.
CRITICAL: It will take several hours to complete. Do not close the terminal window until it finishes.Note: The exact duration of this step primarily depends on the user’s computational processing power and internet connection speed.Note: ‘rumball = "docker run --rm -it --user $(id -u):$(id -g) -v $(pwd):/work rnakato/rumball"’ sets up an alias called ‘rumball’ that will run a Docker container based on the image ‘rnakato/rumball.’ The options ‘--rm’ and ‘-it’ ensure that the container is removed after exiting and that it runs interactively, respectively. The option ‘--user $(id -u):$(id -g)’ specifies the user and groups IDs to use inside the container, ensuring that the container runs with the same user/group permissions as the host user. The option ‘-v $(pwd):/work’ mounts the current working directory on the host machine ‘($(pwd))’ to the ‘/work’ directory inside the container. This allows data to be shared between the host and the container. The command ‘mkdir -p fastq’ creates a new directory named ‘fastq’ if it does not already exist (with ‘-p’). The command ‘$rumball fasterq-dump SRR710092 -O fastq -v’ executes the ‘fasterq-dump’ command from the SRA-Toolkit inside the RumBall Docker container to download the SRA file with ID SRR710092 into the directory fastq (with ‘-O’). Fasterq-dump assumes each SRA contain paired-end reads (‘--split-files’) by default. To improve the speed of file conversion, the parameter ‘-e’ followed by number of threads can allocate more CPU cores to the task and ‘-p’ (progress) provides more detailed information upon completion. You can assess the number of cores in your system by typing ‘nproc --all’ on your Linux terminal. Pause point: You will notice that the last four lines are essentially repetitions, with only the SRA sample IDs being replaced. Alternatively, as a best-practice, we recommend users to create an external file containing the list of the SRA IDs to be downloaded and the sample information, and group to facilitate and automate further steps.
Pause point: You will notice that the last four lines are essentially repetitions, with only the SRA sample IDs being replaced. Alternatively, as a best-practice, we recommend users to create an external file containing the list of the SRA IDs to be downloaded and the sample information, and group to facilitate and automate further steps. -
b.Create sample_info.csv file:>cat >sample_info.csv <<EOF>SRR710092,HEK293_Control_rep1,control>SRR710093,HEK293_Control_rep2,control>SRR710094,HEK293_siCTCF_rep1,siCTCF>SRR710095,HEK293_siCTCF_rep2,siCTCF>EOFNote: This file structure has been described in step 2e.
-
c.Run the code below to have fasterq-dump automatically downloading all samples you have included in the ‘sample_info.csv’ file.>rumball="docker run --rm -it --user root -v $(pwd):/work rnakato/rumball">mkdir -p fastq>cat sample_info.csv|while IFS=, read -r id name group; do>$rumball prefetch "$id" -O /work>$rumball fasterq-dump "$id" -O /work/fastq -e 16 -p>$rumball pigz -p 16 "./fastq/${id}_1.fastq">$rumball pigz -p 16 "./fastq/${id}_2.fastq">doneNote: a while loop is used to process each line in ‘sample_info.csv.’ The ‘IFS = , read -r id name group’ command is a crucial part of this loop. ‘IFS = ,’ sets the internal field separator to a comma, which is useful for parsing CSV files. ‘read -r id name group’ reads each line, splitting it at the comma. The ‘id’ variable captures the first field (the SRA ID), similarly to ‘name’ and ‘group’ columns. Once the SRA ID is extracted, the script executes a series of commands for each line. First, ‘prefetch’ is used to download the dataset from the SRA ID, followed by ‘fasterq-dump’ for converting the SRA data into FASTQ format. After the conversion, ‘pigz’ is used to compress the files into gz format, utilizing multiple threads as defined by the ‘-p’ option (in this case, we use 16 CPUs). Compression is recommended as these files often contain several GBs of size.Note: ‘--user root’ is required to allow fasterq-dump to create temporary files in the image.
 Pause point: The outputs of the step 3c are eight FASTQ files tagged as ‘.fastq.gz’ in fastq directory.
Pause point: The outputs of the step 3c are eight FASTQ files tagged as ‘.fastq.gz’ in fastq directory.
-
a.
-
4.
Obtaining the reference genome.
A reference genome and its corresponding gene annotation are essential for RNA-seq analysis. RumBall provides the scripts ‘download_genomedata.sh’ and ‘build-index-RNAseq.sh’ to download the data and build the index files, respectively.-
a.Download the reference genome and annotation:>rumball="docker run --rm -it --user root -v $(pwd):/work rnakato/rumball">build=GRCh38>Ddir=Ensembl-$build>mkdir -p log>mkdir -p $Ddir># Download genome and gtf>$rumball download_genomedata.sh $build /work/$Ddir 2>&1 | tee log/Ensembl-$build># make index for STAR-RSEM>$rumball build-index-RNAseq.sh rsem-star $build /work/$DdirNote: ‘build = GRCh38’ sets the variable build to GRCh38, which is a code for the human genome version to use. This establishes the directory naming convention, with ‘Ensembl-GRCh38’ as an example. ‘Ddir = Ensembl-$build/’ formulates a directory path string incorporating the genome build name. ‘mkdir -p log’ creates a ‘log’ directory. This ‘log’ directory will store relevant logs such as errors or process statuses. ‘$rumball download_genomedata.sh $build $Ddir 2>&1 | tee log/Ensembl-$build’ runs the ‘download_genomedata.sh’ script to download the genome and annotation data, storing in ‘$Ddir’ and capturing the logs. ‘$rumball build-index-RNAseq.sh rsem-star $build $Ddir’ executes the build-index.sh script to generate index files from the reference data in ‘$Ddir,’ which will be used by mapping tools in the next step.Note: Every piece of information placed to the right side of a ‘#’ is called a ‘comment’ and is not processed by the terminal. This is useful for incorporating instructions or additional documentation directly into the code.Note: To know which organisms are currently available in RumBall pipeline, type:>docker run --rm -it --user $(id -u):$(id -g) -v $(pwd):/work rnakato/rumball download_genomedata.sh
-
b.Check files in the current directory:>ls
 Pause point: Successfully completing step 4 will generate three new directories: ‘Ensembl-GRCh38,’ which contains the genome with its annotation file and the newly built indexes for mapping tools; ‘fastq,’ housing the FASTQ files; and the ‘log’ directory, where users can monitor errors.
Pause point: Successfully completing step 4 will generate three new directories: ‘Ensembl-GRCh38,’ which contains the genome with its annotation file and the newly built indexes for mapping tools; ‘fastq,’ housing the FASTQ files; and the ‘log’ directory, where users can monitor errors.
-
a.
Check strandedness
Timing: 23 min
There are two types of library preparation protocols for RNA-seq analysis based on mRNA library template preparation: stranded and non-stranded RNAs.29 In non-stranded RNAs, the proportion of reads sequenced from both the forward and reverse strands is similar. However, in stranded RNAs, most reads come from the reverse strand. This distinction is crucial for downstream analysis and is typically provided on the GEO profile page.
Note: If the strandedness of RNA-seq data is uncertain, you can verify it using the script check_stranded.sh as shown below:
>rumball="docker run --rm -it --user $(id -u):$(id -g) -v $(pwd):/work rnakato/rumball"
>cat sample_info.csv | while IFS=, read -r id name group; do
>echo $name
>fq1="/work/fastq/${id}_1.fastq.gz"
>$rumball check_stranded.sh human "$fq1"
>done
Note: The command ‘echo $name’ displays the name of the sample currently being processed, helping to track the script's progress. ‘fq1 = /work/fastq/${id}_1.fastq.gz’ constructs a file path string for the first FASTQ file corresponding to the SRA ID currently being processed. This file is expected to be located in the directory ‘fastq.’ Finally, ‘$rumball check_stranded.sh human $fq1’ checks the strandedness of the RNA-Seq data, using ‘human' as a reference and ‘$fq1’ as the input FASTQ file.
Pause point: The output of this command exhibits the counts for each strand for each sample on the screen. An example is provided below.
Note: In the datasets we examined, most reads were mapped to the minus strand (denoted as ’-’), indicating that this RNA-seq data is stranded.
Reported 27787970 alignments
540264 +
27247706 -
Mapping reads by STAR and estimating gene expression by RSEM
Timing: 4.5 h
RumBall supports the most popular mapping tools: STAR,14 Bowtie2,13 kallisto,17 and salmon.15 In this tutorial, we use STAR as an example. STAR is one of the fastest and the most accurate FASTQ read alignment tools.30 While kallisto and salmon also estimate the gene expression levels, STAR and bowtie2 do not. Therefore, RumBall provides useful scripts to systematically perform the gene expression estimation with RSEM18 from the output of STAR.
-
5.
The following commands run STAR for all samples:
>rumball="docker run --rm -it --user root -v $(pwd):/work rnakato/rumball"
>build=GRCh38
>Ddir=Ensembl-$build
>mkdir -p log
>cat sample_info.csv | while IFS=, read -r id name group; do
> echo "$name"
> fq1="/work/fastq/${id}_1.fastq.gz"
> fq2="/work/fastq/${id}_2.fastq.gz"
$rumball star.sh -d /work/star paired "$name" "$fq1 $fq2" $Ddir reverse > "log/$name.star.sh"
>done
Note: This process assigns each pair of reads to ‘fq1’ and ‘fq2’ from each sample and runs our ‘star.sh’ script. ‘$rumball star.sh -d /work/star paired $name “$fq1 $fq2” $Ddir reverse > log/$name.star.sh’ performs alignment of the RNA-Seq reads using STAR. Within this command: ‘-d’ is for the output directory, ‘paired’ specifies the paired-end type, ‘$name’ provides custom names for each sample, input fastq files are represented by ‘$fq1 $fq2,’ the directory with genome data is indicated with ‘$Ddir,’ and the strandedness of the RNA-Seq data can be denoted as ‘reverse’, ‘forward’ or ‘none’. The logs are stored for each sample using ‘> log/$name.star.sh’ and provides detailed information of the mapping progress.
-
6.
Then, quantify the reads with RSEM:
>rumball="docker run --rm -it --user $(id -u):$(id -g) -v $(pwd):/work rnakato/rumball"
>build=GRCh38
>Ddir=Ensembl-$build
>Ctrl=""
>siCTCF=""
>while IFS=, read -r id name group; do
> if [ "$group" = "control" ]; then
> Ctrl+="/work/star/$name "
> elif [ "$group" = "siCTCF" ]; then
> siCTCF+="/work/star/$name "
> fi
>done < sample_info.csv
>mkdir -p Matrix_deseq2
>$rumball rsem_merge.sh "$Ctrl $siCTCF" /work/Matrix_deseq2/HEK293 $Ddir
Note: ‘Ctrl’ and ‘siCTCF’ are variables that store the locations of STAR-aligned output for the Control and siCTCF samples, respectively. We first make sure they are empty by assigning “” to each variable. Then we obtain the sample names for each control group storing the absolute path. Here we use the third column of sample_info.csv file. Please adapt accordingly when using different names. ‘mkdir -p Matrix_deseq2’ creates a directory named Matrix_deseq2 for storing the output files from RSEM and DESeq2 differential expression analysis. ‘rsem_merge.sh’ script merges the generated expression data for both the Control and siCTCF samples into a single matrix, which is saved in ‘Matrix_deseq2/HEK293.’ Indicate the mapping reference location via ‘$Ddir.’
Differential gene expression analysis and gene ontology analysis
Timing: 15 min
Differential gene expression analysis using RNA-seq is essential for identifying genes with varying expression under different conditions, providing critical insights into biological and pathological processes. We utilize DESeq2, a R package that employs a model based on the negative binomial distribution. This model adjusts for sample variations and provides robust statistical inference for identifying differentially expressed genes (DEGs) between two groups. The results are adjusted using the Benjamini-Hochberg method with an FDR cutoff < 0.01.20 After identifying upregulated and downregulated DEGs based on positive and negative ‘log2FoldChange’ values, we proceed to determine the enriched gene ontologies associated with the top-ranked genes using ClusterProfiler. Log2FoldChange is a commonly used metric to represent the range of expression changes between two conditions. A log2FoldChange of 1 signifies that gene expression is 2-fold higher (or lower) in the second.
-
7.
Run the DESeq2 script:
>rumball="docker run --rm -it --user $(id -u):$(id -g) -v $(pwd):/work rnakato/rumball"
>$rumball DESeq2.sh -l 1 –c 2 -t 0.05 -n 500 Matrix_deseq2/HEK293 2:2 control:siCTCF Human
Note: The DESeq2.sh script performs differential gene expression analysis using DESeq2 and gene ontology enrichment with ClusterProfiler.24 The script’s arguments specify the input matrix location (‘Matrix_deseq2/HEK293’), the number of samples in each group (‘2:2’), the names of the groups (‘control:siCTCF’), and the species (‘Human’). It’s important to use the same nomenclatures as those in ‘sample_info.csv’ file for the groups, and comparisons should be conducted in a similar manner. The ‘-l 1’ option defines the log2FoldChange threshold and ‘-c 2’ allows switching between annotations provided (in our tutorial, ‘1’ for Ensembl ID and ‘2’ for gene symbol). For GO enrichment, the ‘enrichGO’ function and a list of Human genes from ‘org.Hs.eg.db’ are used to perform the hypergeometric test, which is corrected by the Benjamini-Hochberg method. A p-value cutoff of <0.05 is defined by the ‘-t’ parameter. The default value for top-ranked genes is set at 500, but this can be adjusted using the ‘-n’ option. Results are stored in the same directory as the DEG analysis.
Note: More information on the species and parameters can be obtained using the command:
>docker run --rm -it --user $(id -u):$(id -g) -v $(pwd):/work rnakato/rumball DESeq2.sh
-
8.
To observe the variance in gene expression among samples, users can review the PCA and hierarchical clustering plots (Figures 1 and 2). Open the files with suffixes ‘.samplePCA.pdf’ and ‘.sampleClustering.pdf’ for gene and isoforms, respectively.
-
9.
Similarly, to investigate the variance in the gene expression values and the fitted dispersion calculated by DESeq2, users should inspect the ‘.Dispersionplot.pdf’ and ‘.MAplot.pdf’ plots (Figures 3 and 4).
-
10.
The most expressed genes, which significantly influence our experiment, can be identified through the '.HighlyExpressedGenes.pdf’ plot (Figure 5).
Pause point: By reaching this point, you have successfully performed a complete DEG analysis. The results of differential gene expression analysis and overall gene expression values are contained under the files ending with ‘.DEGs.tsv’ at the Matrix_deseq2 directory. For a quickly access to up- and down-regulated DEGs, we provided the lists in separated files.
-
11.
To access the content of TSV files using the Bash ‘head’ command, type (output shown in Figure 6):
>head -n 20 ∗DEGs.tsv
Note: Alternatively, user can open the .tsv file using Microsoft Excel or similar software (visualization example in Figure 7).
Figure 1.
PCA plot showing sample variance
The relative positioning of the points on the plot provides insights into the similarities and differences between samples, with closer points indicating more similar gene expression profiles. PCA plots assist in spotting outliers, batch effects, and clustering patterns related to biological or experimental conditions. In this plot, PC1 (x-axis) captures the most significant variance in the data, often reflecting major sources of variation such as dominant biological differences or experimental conditions. PC2 (y-axis) represents the second-most variance, orthogonal to PC1, and can reveal additional, more subtle variations in the dataset. Together, PC1 and PC2, offer a comprehensive view of the overall structure and variability in the gene expression data.
Figure 2.
Hierarchical clustering of gene expression data across conditions
Hierarchical clustering can group genes and samples with similar expression patterns, aiding in identifying co-expressed gene groups or similar sample profiles. The heatmap provides a compact graphical representation of large gene expression datasets, emphasizing patterns and relationships in the data. The clustering of similar conditions is expected.
Figure 3.
Dispersion plot of the genes after model-fitting
A dispersion plot in DESeq2 depicts gene-wise dispersion values against mean normalized counts. The x-axis represents the mean normalized counts, and the y-axis shows the dispersion (variance/mean). Fitted dispersion values are plotted as a line, capturing the expected trend. Deviation from this line indicates genes with unexpected variability across replicates. Such a plot is instrumental in visualizing and accounting for both biological and technical variability inherent in RNA-seq data.
Figure 4.
MA plot of differential gene expression
Points above and below a horizontal line at y = 0 on the plot indicate genes that are upregulated and downregulated, respectively. Significantly differential genes are highlighted in blue. "shrunken apeglm" removes the high variance of low expression genes. The MA plot provides a visual summary of the differential expression results, helping to identify patterns and potential outliers.
Figure 5.
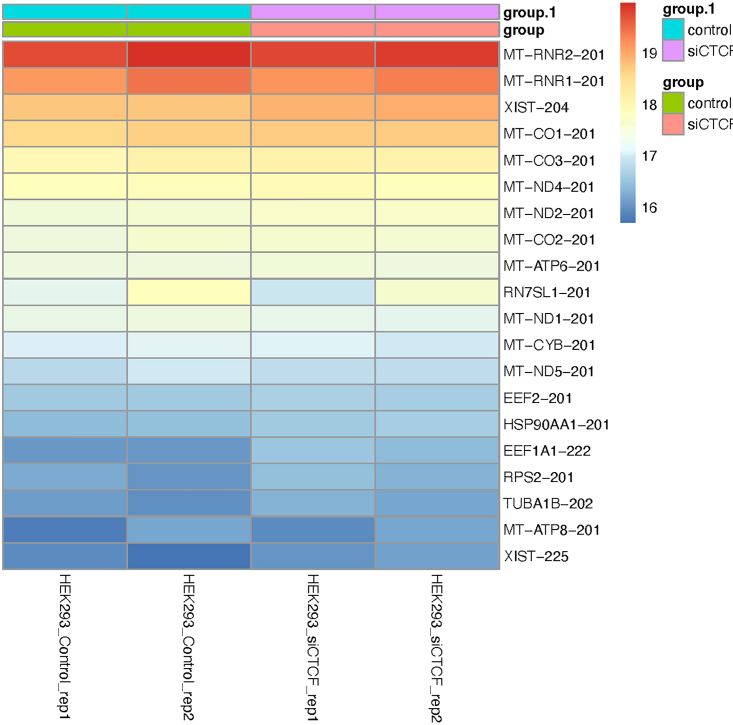
Heatmap of highly expression levels across conditions
The color scale represents normalized isoforms expression levels, with red indicating high expression and blue indicating low expression. Each row corresponds to a specific isoform, and each column represents a sample from a different experimental condition.
Figure 6.
Example of file visualization using command line
Only the first n = 20 rows are shown. Alternative, the command tail shows from the opposite direction.
Figure 7.
List of differential gene expression using Microsoft Excel
Same table as Figure 6 open in Microsoft Excel.
The command ‘wc’ (word count) can be applied to access the number of DEGs in each file. Parameter ‘-l’ stands for lines and show counts of entries in each file (Figure 8).
>wc -l ∗DEGs.tsv
-
12.
To check the significant differentially expressed genes, user can check the ‘Volcano.pdf’ plot (Figure 9).
-
13.
The gene ontology enrichment analysis produces separate plots for up- and down-DEGs that are shown in ‘DEGs_top500.pdf’ files (Figure 10).
Figure 8.
Number of DEGs contained in each of the files
Terminal screen with quick inspection of number of genes in each file (including first line as header).
Figure 9.
Volcano plot and top differentially expressed genes
Points far from the vertical x = 0 line indicate high fold changes, while those higher on the plot suggest greater significance. Genes of interest, determined by fold change thresholds and significance levels, are highlighted in the plot’s upper extremes.
Figure 10.
Gene Ontology enrichment analysis highlighting functional categories significantly represented from the list of down-regulated genes
This analysis provides insights into potential biological process affected in HEK293 knock-down cells. Enrichment significance was determined using a hypergeometric test with p-values adjusted by the Benjamini-Hochberg method (cutoff < 0.05). The top 500 down regulated genes were considered for this analysis.
Expected outcomes
The RumBall pipeline, designed for RNA-seq data analysis, effectively organizes its outputs into multiple directories to ensure systematic organization throughout the various stages of analysis. This organization facilitates a comprehensive suite of outputs, initiating with STAR mapping results that encompass aligned reads to both the genome and transcriptomes, alongside gene and transcript expression data, complemented by detailed mapping statistics. Following this, the pipeline integrates gene expression values, delivering both raw counts and TPM-normalized data, which culminates in consolidated expression files. Differential expression analysis conducted via DESeq2 generates an extensive array of outputs, ranging from a complete gene list to specific lists that detail differentially expressed genes (DEGs), both upregulated and downregulated, alongside a variety of graphical representations such as dispersion, MA, volcano plots, and heatmaps for nuanced data interpretation. Additionally, gene ontology enrichment analysis performed by ClusterProfiler introduces an additional dimension to the analysis, offering insights into the top DEGs and the biological mechanisms they signify.
Limitations
The RumBall pipeline is designed to integrate functionality, reproducibility, and require minimal modifications. Despite its designed efficiencies, the pipeline has certain limitations that potential users need to consider. Firstly, a significant limitation is the absence of batch correction. The pipeline assumes that all samples used are directly comparable, either originating from identical laboratory conditions or produced in similar environments. Therefore, incorporating covariates may not yield realistic DEGs, which is a crucial consideration for users. Secondly, the pipeline effectiveness in gene ontology analysis is inherently tied to the size of the DEG list it generates. Smaller lists might lead to less reliable outputs or may fail to obtain any enrichment, whereas excessively long lists (e.g., over 1000 genes) might not accurately reflect the underlying biological phenomena due to overrepresentation. Lastly, the pipeline present customization limitations for advanced users. Those seeking to tailor de pipeline for specific needs or to incorporate particular modifications may find themselves needing to rebuild the Docker image. Such a task demands proficiency in Docker and pipeline configuration, which may not be feasible for all users.
Troubleshooting
Problem 1
Tried to pull RumBall from DockerHub with permission denied error at Obtaining RumBall.
Potential solution
Include ‘sudo’ before the command.
>sudo docker pull rnakato/rumball
Alternatively, the system administrator can create a new group and add users, allowing them to use Docker without sudo described at 6. Create the docker group section. Detailed information can be obtained at https://docs.docker.com/engine/install/linux-postinstall/.
Problem 2
Error with SRA toolkit configuration at preparing datasets and reference genome.
Potential solution
-
•
Run the command:
> vdb-config --interactive
-
•
Select ‘Enable Remote Access.’
-
•
Type ‘S’ followed by ‘Enter’ to confirm.
-
•
Type ‘X’ followed by ‘Enter’ to leave.
-
•
Rerun the command.
Problem 3
Error: The sample list does not match the IDs during the step preparing datasets and reference genome.
Potential solution
Make sure that the number of IDs and names matches. Check whether the order of the names also corresponds to the sample IDs.
Problem 4
Issues with duplicate gene names in DESeq2 analysis. When using gene symbols as identifiers in DESeq2, users may encounter errors or unexpected results due to the presence of duplicated gene names. DESeq2 requires unique identifiers for each gene, and gene symbols can often map to multiple transcripts or gene variants, leading to duplicates.
Potential solution
In this tutorial, we use Gene Symbols for better interpretation of the results. However, if users are interested in detailed expression of transcript isoforms we recommend using Ensembl IDs, which are unique for each transcript and avoid the issue of duplication. Users experiencing problems with gene symbols can switch back to Ensembl IDs by modifying the annotation column in our tool’s settings (choose either column 1 or 2 for Ensembl IDs).
Problem 5
ClusterProfiler fails to generate enrichment plots. Users may encounter a situation where ClusterProfiler, a tool used for statistical analysis and visualization of functional profiles for genes and gene clusters, does not generate the expected enrichment plots. This issue often arises when the number of genes provided as input, specifically those identified as up-regulated or down-regulated, is too low. An insufficient number of genes can lead to a scenario where ClusterProfiler is unable to find significant enrichment, resulting in the absence of a plot.
Potential solution
Users should consider increasing the number of genes used as input for the enrichment analysis. This can be achieved by adjusting the parameter -n in DESeq2.sh script. Increasing the value of -n allows for a larger set of genes to be considered, thereby enhancing the likelihood of identifying significant enrichments. It’s important to balance the number of input genes; too many genes might lead to diluted or less specific results, whereas too few may not capture enough information for meaningful analysis.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Ryuichiro Nakato (rnakato@iqb.u-tokyo.ac.jp).
Technical contact
For tutorial and technical issues contact Luis Nagai (nagai@iqb.u-tokyo.ac.jp).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Data used in this tutorial has been deposited and is available at GEO, under the accession number GEO: GSE44267 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44267). Codes and scripts are accessible on GitHub (https://github.com/rnakato/RumBall) and Zenodo: (https://doi.org/10.5281/zenodo.10614707). A comprehensive and constantly updated manual and official documentation can be accessed at the RumBall web manual page https://rumball.readthedocs.io/en/latest/index.html. Additionally, the RumBall Docker image is available at DockerHub: (https://hub.docker.com/r/rnakato/rumball).
Acknowledgments
We thank all lab members for the support and discussion, as well as users that tested and commented on our GitHub page. This work was supported by the Japan Agency for Medical Research and Development under grant number JP23gm6310012h0004.
Author contributions
L.A.E.N. and R.N. created this protocol. L.A.E.N. performed the analyses and first tested the scripts. S.L. tested the protocol and suggested improvements. R.N. conceived the project. All authors read and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Luis Augusto Eijy Nagai, Email: nagai@iqb.u-tokyo.ac.jp.
Ryuichiro Nakato, Email: rnakato@iqb.u-tokyo.ac.jp.
References
- 1.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 2.Di Tommaso P., Chatzou M., Floden E.W., Barja P.P., Palumbo E., Notredame C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017;35:316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- 3.Köster J., Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 4.Blankenberg D., Kuster G.V., Coraor N., Ananda G., Lazarus R., Mangan M., Nekrutenko A., Taylor J. Galaxy: A Web-Based Genome Analysis Tool for Experimentalists. Curr. Protoc. Mol. Biol. 2010;89 doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etoh K., Nakao M. A web-based integrative transcriptome analysis, RNAseqChef, uncovers the cell/tissue type-dependent action of sulforaphane. J. Biol. Chem. 2023;299 doi: 10.1016/j.jbc.2023.104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge S.X., Son E.W., Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinf. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harshbarger J., Kratz A., Carninci P. DEIVA: a web application for interactive visual analysis of differential gene expression profiles. BMC Genom. 2017;18:47. doi: 10.1186/s12864-016-3396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson J.W., Sklenar J., Barnes A.P., Minnier J. The START App: a web-based RNAseq analysis and visualization resource. Bioinformatics. 2017;33:447–449. doi: 10.1093/bioinformatics/btw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merkel D. Docker: lightweight Linux containers for consistent development and deployment. Linux J. 2014;2 [Google Scholar]
- 10.Zuin J., Dixon J.R., van der Reijden M.I.J.A., Ye Z., Kolovos P., Brouwer R.W.W., van de Corput M.P.C., van de Werken H.J.G., Knoch T.A., van IJcken W.F.J., et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. USA. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 18.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR : a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frazee A.C., Pertea G., Jaffe A.E., Langmead B., Salzberg S.L., Leek J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 24.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolberg L., Raudvere U., Kuzmin I., Vilo J., Peterson H. gprofiler2 -- an R package for gene list functional enrichment analysis and namespace conversion toolset g. F1000Res. 2020;9:709. doi: 10.12688/f1000research.24956.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database Collaboration The Sequence Read Archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin J.Z., Yassour M., Adiconis X., Nusbaum C., Thompson D.A., Friedman N., Gnirke A., Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D.J., Elo L.L., Zhang X., Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data used in this tutorial has been deposited and is available at GEO, under the accession number GEO: GSE44267 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44267). Codes and scripts are accessible on GitHub (https://github.com/rnakato/RumBall) and Zenodo: (https://doi.org/10.5281/zenodo.10614707). A comprehensive and constantly updated manual and official documentation can be accessed at the RumBall web manual page https://rumball.readthedocs.io/en/latest/index.html. Additionally, the RumBall Docker image is available at DockerHub: (https://hub.docker.com/r/rnakato/rumball).