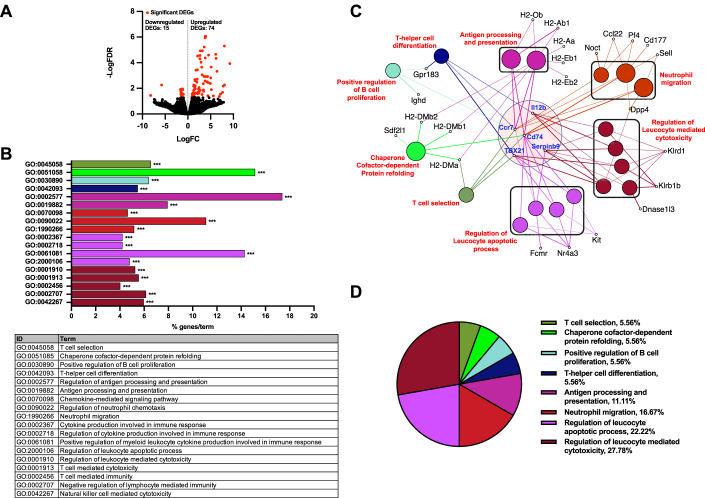
Figure 5. RNA-seq analyses of CD64+MHCII+ and CD64+MHCII- macrophages reveal differentially expressed genes that influence multiple immunological aspects.
(A) CD64+MHCII+ and CD64+MHCII- macrophages were sorted from CHIKV-infected joints (n = 5 animals per group) at 6 dpi and processed for RNA-seq. A total of 89 DEGs were identified. (B) Representative gene ontology terms and pathways are depicted, including genes involved in Th1 and Th2 differentiation as well as antigen processing and presentation. Analysis was carried out with a two-sided hypergeometric test, corrected with Bonferroni stepdown. ***P = < 0.001 (refer to Dataset EV1 for exact P values). (C) Gene ontology terms are presented as nodes and clustered together based on the similarity of genes present in each term or pathway. Ccr7, Cd74, Il12b, TBX21, and Serpinb9 are identified as genes highly associated with many of these terms. (D) A functionally grouped network of enriched pathways or terms based on the 89 DEGs were generated by querying the Gene Ontology - biological database with ClueGo. Source data are available online for this figure.