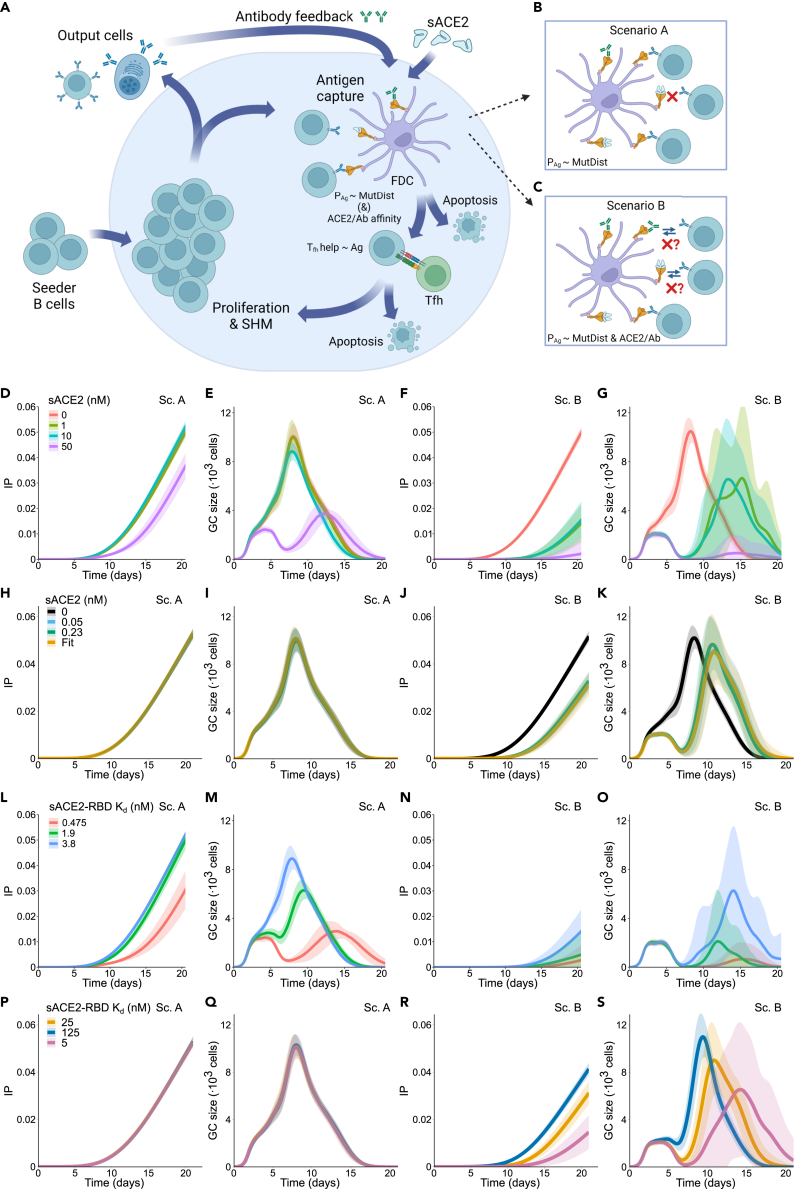
Figure 4.
Soluble ACE2 interferes with the germinal center reaction in silico
(A) Scheme depicting the germinal center (GC) response: GC reaction starts with an influx of low-affinity seeder cells. These cells are in a pro-apoptotic state and must acquire survival signals by antigen acquisition and T follicular helper cell (Tfh cell) signaling. Ag binding probability (PAg) is proportional to the mutational distance of the BCR to the Ag (MutDist, see STAR Methods and supplemental information). The Tfh cells preferentially provide signal to B cells that acquire larger amounts of antigen. Some of the selected B cells exit the GC and become output cells, whereas the rest recycle back and proliferate with mutations, resulting in daughter cells of higher affinity. These new cells again compete for survival signals, resulting in progressive B cell affinity increase. Serum sACE2 and the antibodies produced by output B cells can feedback to the GC by making antigen acquisition harder (mechanisms in B and C).
(B) Epitope masking mechanism (scenario A): sACE2 and antibodies can bind to the antigen and mask it, thus lowering the amount of antigen available for B cells.
(C) Epitope masking and mean field Ab-B cell competition mechanism (scenario B): BCR probability of binding the unmasked Ag is reduced depending on the average affinity of sACE2 and endogenous Abs (ACE2/Ab affinity).
(D–K) Simulation results for different sACE2 concentrations (0, 1, 10, and 50 nM constant concentration in D–G; 0, 0.05, 0.23 nM constant concentration and 0.05–0.23 nM linear increase in H–K): IP (D, H) and GC size (E, I) with scenario A; IP (F, J) and GC size (G, K) with mean field Ab-B cell competition. (L–S) Simulation results for different sACE2 affinities (0.475, 1.9, and 3.8 nM in L–O; 5, 25, 125 nM in P–S): IP (L, P) and GC size (M, Q) with epitope masking only; IP (N, R) and GC size (O, S) with scenario B. Mean (continuous lines) and standard deviation (shaded area) of simulations for a total of 20 simulated GCs are shown. Schemes (A–C) are created with BioRender.