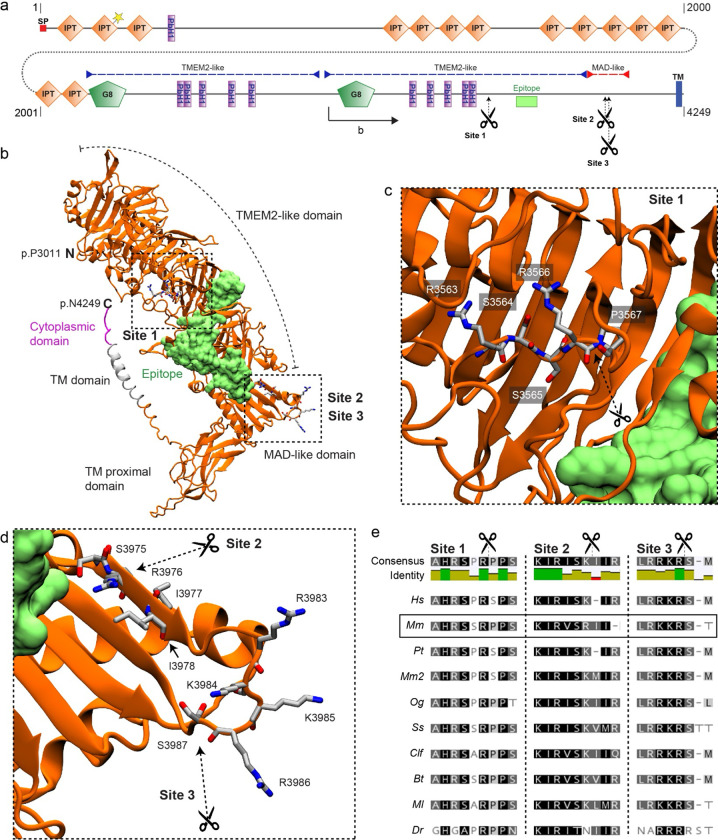
Figure 4. PKHD1L1 contains potential proteolytic cleavage sites.
a, PKHD1L1 domain diagram, based on consensus of UniProt and SMART predictions (domain predictions in Sup. Table 2). Residue numbering includes the signal peptide (20 amino acids) using the mouse reference sequence NP_619615.2. Position of frame shift and premature stop codon in PKHD1L1-deficient mice is depicted by dotted line and yellow star. Purple dashed lines indicate TMEM2-like domains, red dashed line indicates MAD-like domain. The epitope for anti-PKHD1L1 antibody used in this study (NBP2–13765) is depicted by a lime box. Potential cleavage sites at p.R3566 (site 1), p.R3976 (site 2), and p.R3986 (site 3) are depicted with scissors. b, AlphaFold2 modelling was used to predict the structure of a Mm PKHD1L1 protein fragment comprising the residues p.P3011–4249 (illustrated on panel a by black arrow). The protein fragment is depicted in cartoon representation of the ectodomain fragment (orange), transmembrane domain (grey) and cytoplasmic domain (pink). The epitope for anti-PKHD1L1 antibody NBP2–13765 —against p.3670-p.3739, is depicted in surface representation (lime). Potential cleavage are depicted with scissors. The antibody epitope region and the transmembrane domain are separated by the PKHD1L1 MAD-like domain with predicted cleavage sites 2 and 3, suggesting that labelling of the PKHD1L1 ectodomain might be absent following PKHD1L1 cleavage. c, d, Insets representing zoomed-in views of the putative cleavage sites. Images were rendered with the VMD software. e, Protein sequence alignment of the predicted PKHD1L1 cleavage site regions across 10 different species (see accession codes in Sup. Table 1). Analysis was carried out with the Geneious software. Background colors represent the rate of residue conservation: black indicates high-sequence similarity, while white indicates poor residue similarity or its absence. A consensus sequence is shown for all the species at the top of the panel, with a color -coded identity map to illustrate the level of consensus between the different species.