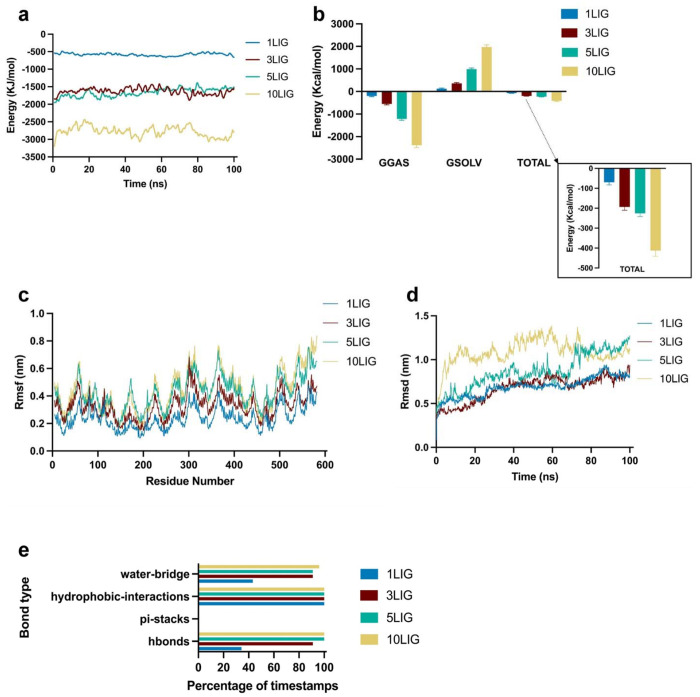
Fig. 4. PtdChos and albumin interaction analysis in all-atoms molecular dynamics simulations.
a, Total linear interaction energies between albumin and various number of ligands systems over simulation time. The total energy represents the sum of Lennard-Jones and coulombic energies. b, Effective free energy components for the different systems. GGAS represents the energy of the gas phase, GSOLV, the energy of solvation, and TOTAL is the sum of the two. Error bars represent mean ±SD. c, Average root mean square fluctuation of albumin residues for the 4 systems. d, Root mean square deviation of PtdChos over time for the 4 types of systems. e, Bond types that are present within each simulation. The y-axis represents the bond types, and the x-axis represents the percentage of simulation timestamps when each type of bond is present.