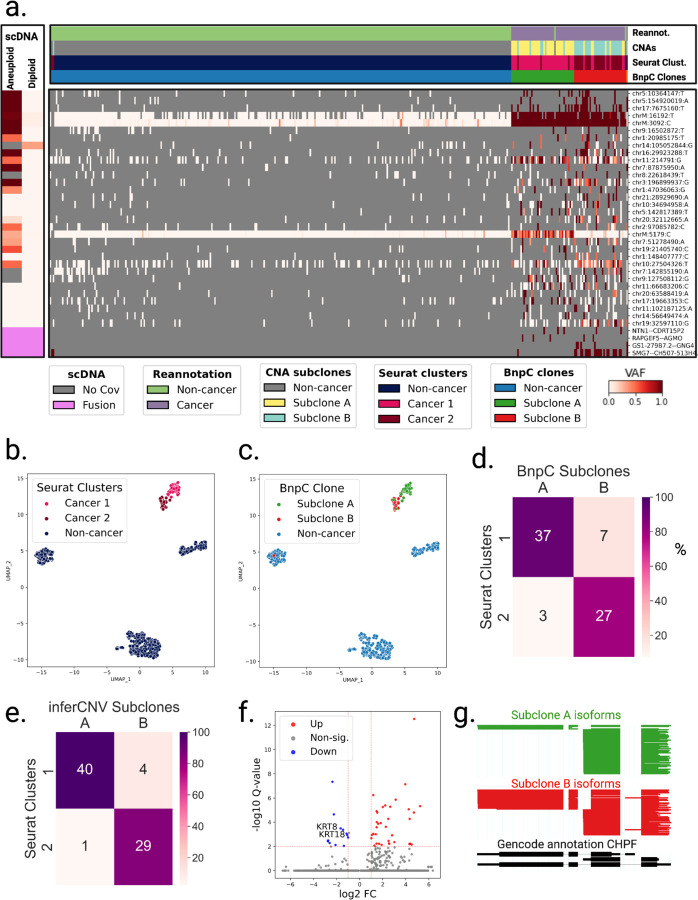
Figure 6: Analysis of intra-tumor heterogeneity using somatic variants detected in LR scRNA-seq in Patient P1.
a. BnpC clustering of single cells from the tumor biopsy of patient P1 (columns) by somatic SNVs and fusions (rows). VAF is depicted as a gradient from white (no mutated reads, VAF=0) to red (only mutated reads, VAF=1). Grey indicates no coverage in the cell at a given locus. Rows are colored by the scDNA VAF of aggregated diploid and aneuploid cells at the loci: SNVs with high aneuploid VAF and low diploid VAF are somatic in scDNA data. Fusions appear in pink. Columns are colored from top to bottom by cell types reannotated by LongSom, CNAs subclones, expression clusters, and BnpC clusters b,c. UMAP embedding of patient P1 gene expression data, colored by (b) cell types reannotated by LongSom and (c) subclones inferred from somatic SNVs and fusions. The dashed line indicates the manual separation between cancer clusters 1 and 2. d,e. Confusion matrix of cells in each expression-derived cancer cluster (rows) and (d) cells in the subclones inferred from BnpC, and (e) cells in the subclones inferred from inferCNV (columns), colored by the percentage of the total number of cells in each subclone and annotated by the absolute numbers. f. Volcano plot of differentially expressed genes identified between subclones B and A. Keratin genes downregulated in subclone B are annotated. g. ScisorWiz representation of CHPF isoforms in subclones A and B. Colored areas are exons, whitespace areas are intronic space, not drawn to scale, and each horizontal line represents a single read colored according to subclones.