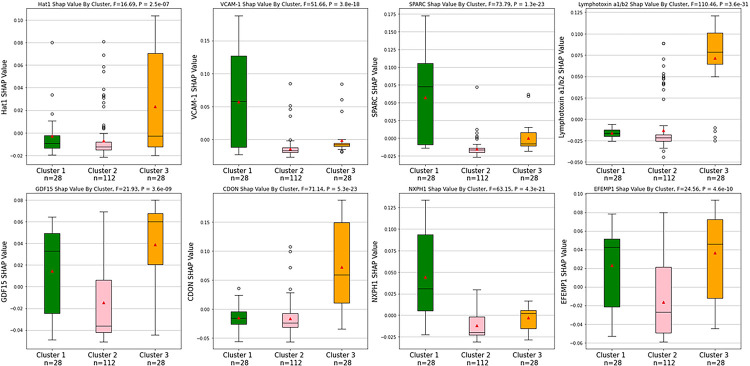
Figure 6. Comparing Feature Selected Biomarker Shapley Additive Explanation Values between Clusters.
Box plots comparing the feature selected biomarkers for predicting the development of low PMA’s SHAP values between the 3 clusters that were illustrated using principal component analysis (PCA) and K-Means clustering. All the biomarkers’ SHAP values were significantly different between the 3 clusters via one-way ANOVA (P < 0.001). Eight feature selected biomarkers for predicting the development of low PMA were used: Histone acetyltransferase type B catalytic subunit (Hat1), Secreted protein acidic and rich in cysteine (SPARC), Lymphotoxin alpha 1/ beta 2 (Lymphotoxin a1/b2), Growth/differentiation factor 15 (GDF15), Cell adhesion molecule-related/down-regulated by oncogenes (CDON), Neurexophilin-1 (NXPH1), Vascular cell adhesion protein 1 (VCAM-1), and EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1). The black lines indicate the medians, the red triangles indicate the means, the circles represent outliers, and the error bars represent 1.5x the interquartile range. There were 168 participants between the 3 groups.