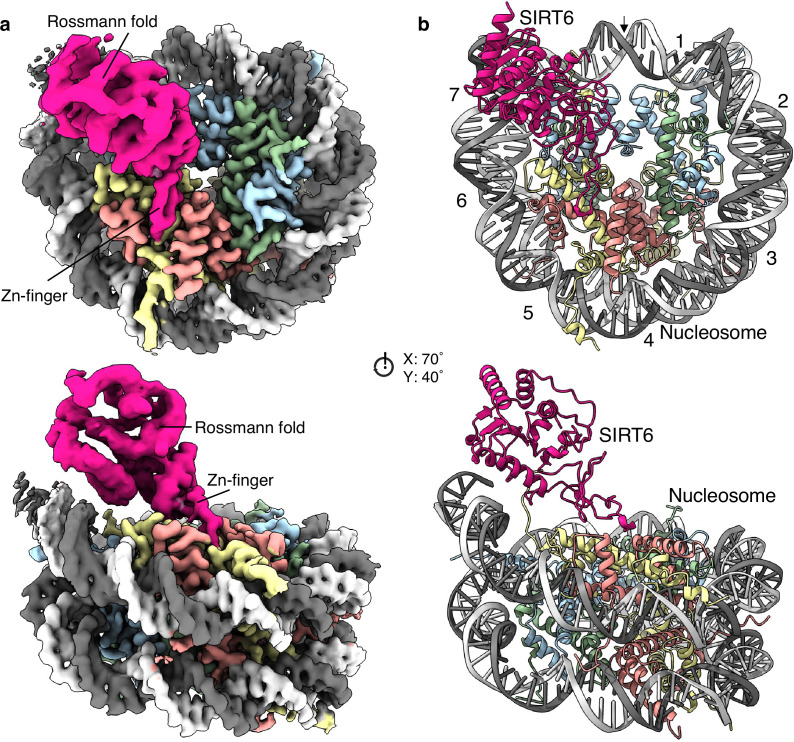
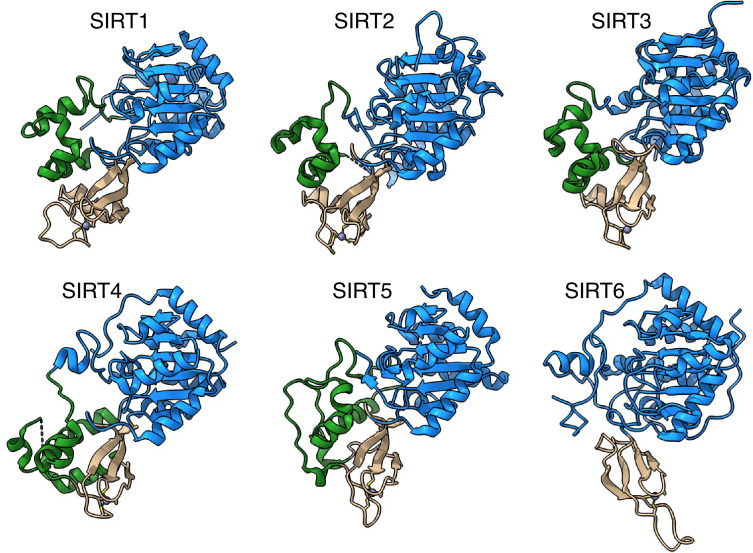
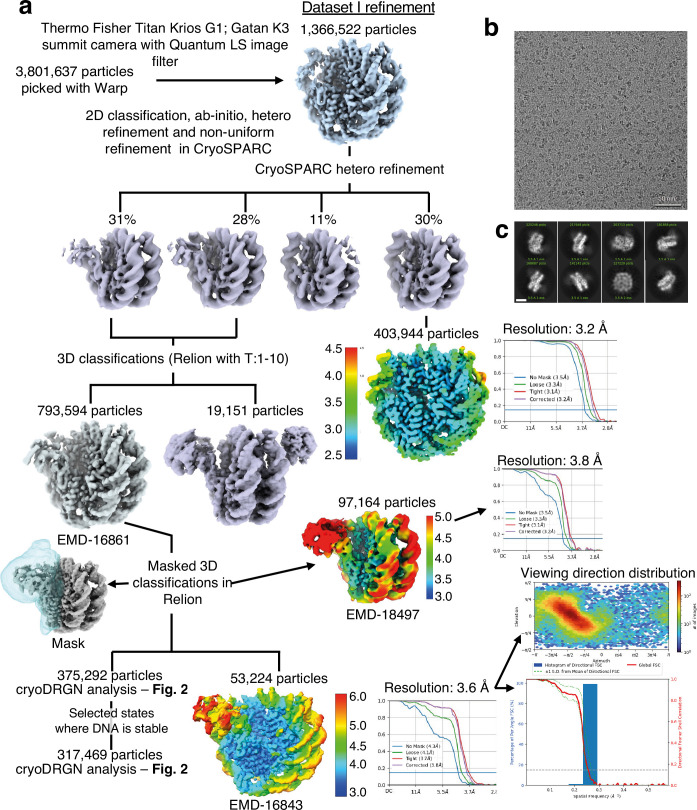
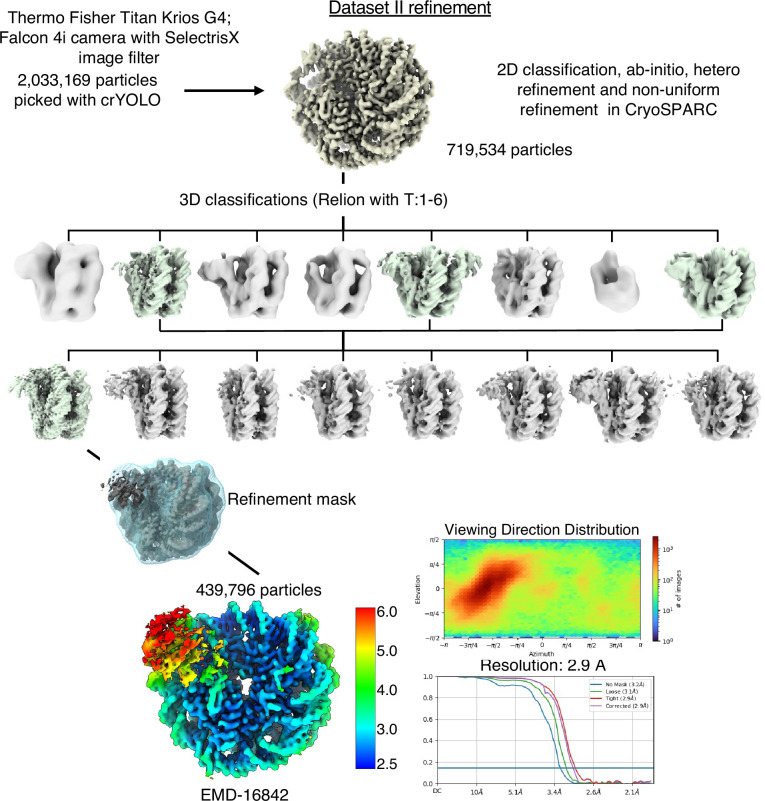
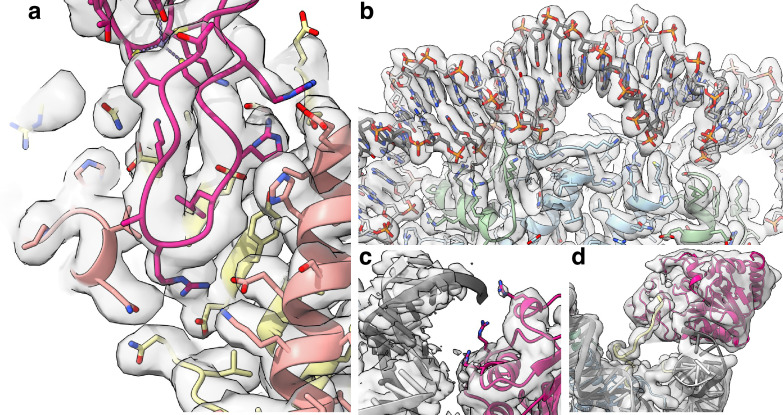
Figure 1. Structure of human SIRT6 in complex with the nucleosome.
(a) Front (top) and side (bottom) views of a composite cryo-EM reconstruction of human SIRT6-nucleosome. Maps from focused refinements of SIRT6 (magenta – Rossmann fold and Zn-finger domains labelled) and the nucleosome (H2A – yellow; H2B – orange; H3 – blue; H4 – green; DNA – light and dim grey). (b) Corresponding views of the atomic model of the complex.