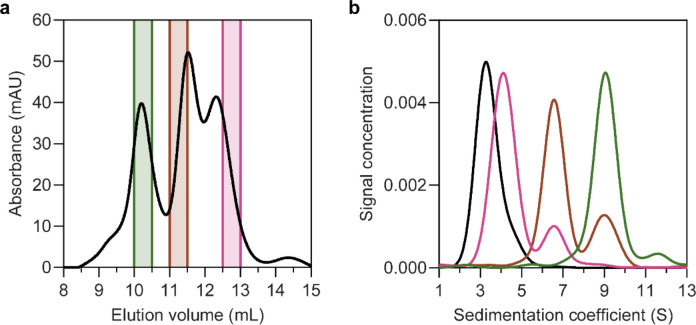
Figure 3. HiSiaQM self-association in lauryl maltose neopentyl glycol (L-MNG) and amphipol.
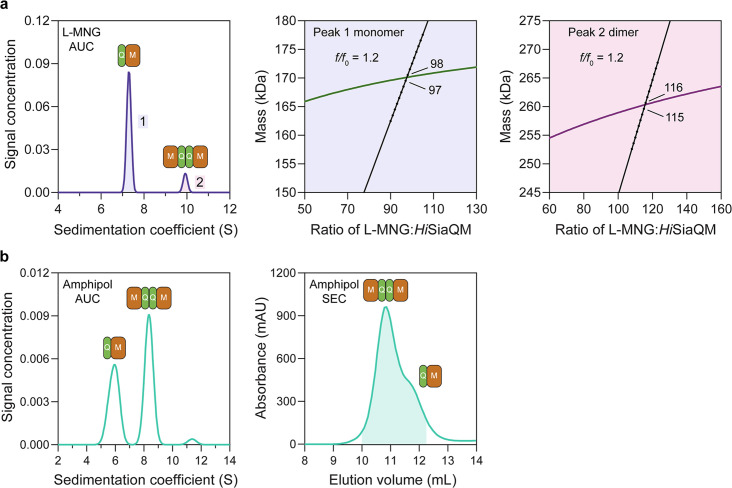
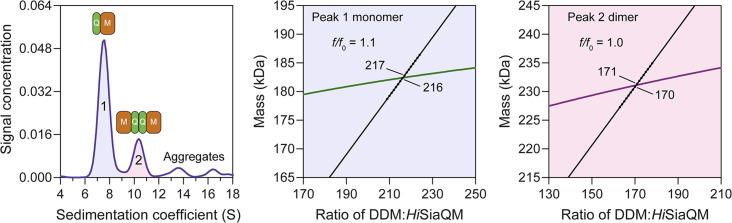
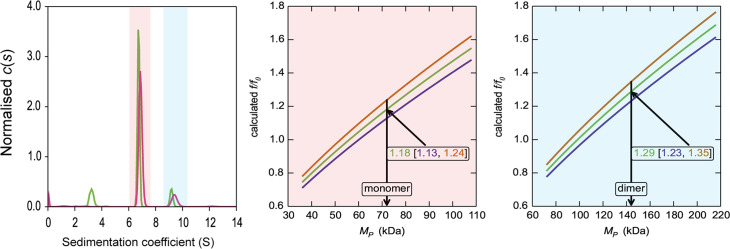
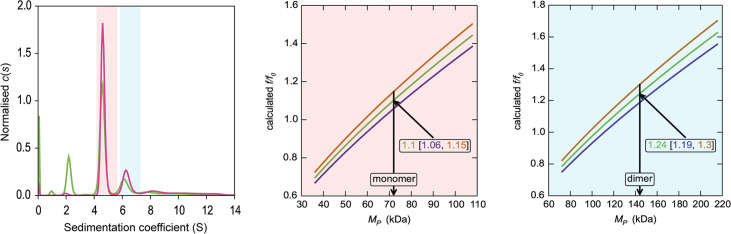
(a) Sedimentation velocity analytical ultracentrifugation (SV-AUC) analysis of HiSiaQM in L-MNG (left panel). Two well-resolved species exist at 7.3S (diffusion coefficient, D = 4.8 × 10–7 cm2/s) and 9.9S (D = 4.2 × 10–7 cm2/s), with the larger peak constituting 85% of the signal. The species at 7.3S (peak 1, blue shading) is most consistent with HiSiaQM as a monomer with ~98 molecules of L-MNG bound (middle panel; green = measured mass, black = theoretical mass), calculated from the experimental sedimentation and diffusion coefficients. These calculations suggest that peak 1 existing as a dimer is unlikely, as the dimeric protein would only have ~14 molecules of L-MNG bound. Additionally, the calculated f/f0 of a monomer for peak 1 is 1.2, consistent with a protein in a detergent micelle. The species at 9.9S (peak 2, pink shading) is most consistent with HiSiaQM as a dimer with ~116 molecules of L-MNG bound (calculated) (right panel; purple = measured mass, black = theoretical mass); peak 2 existing as a monomer is not possible, as the protein clearly has a smaller species in peak 1 and cannot be divided further than a monomer, and a trimer is also unlikely as the trimeric protein would only have ~32 molecules of L-MNG bound (calculated). Additionally, the calculated f/f0 of a dimer for peak 2 is also 1.2, again consistent with a protein in a detergent micelle. These calculations do not account for bound lipid molecules. (b) Left panel: SV-AUC analysis of amphipol-solubilised HiSiaQM (initially purified in L-MNG) shows two distinct species present at 5.9S and 8.3S. These are monomeric and dimeric species as L-MNG-solubilised protein exists as these oligomeric states at 7.3S and 9.9S as in (a). Right panel: representative size-exclusion chromatogram of amphipol-solubilised HiSiaQM favouring the dimeric state. The main peak at ~10.8 mL contains dimeric HiSiaQM and the shoulder at ~11.8 mL contains monomeric HiSiaQM. The sample used for structure determination is shaded turquoise.