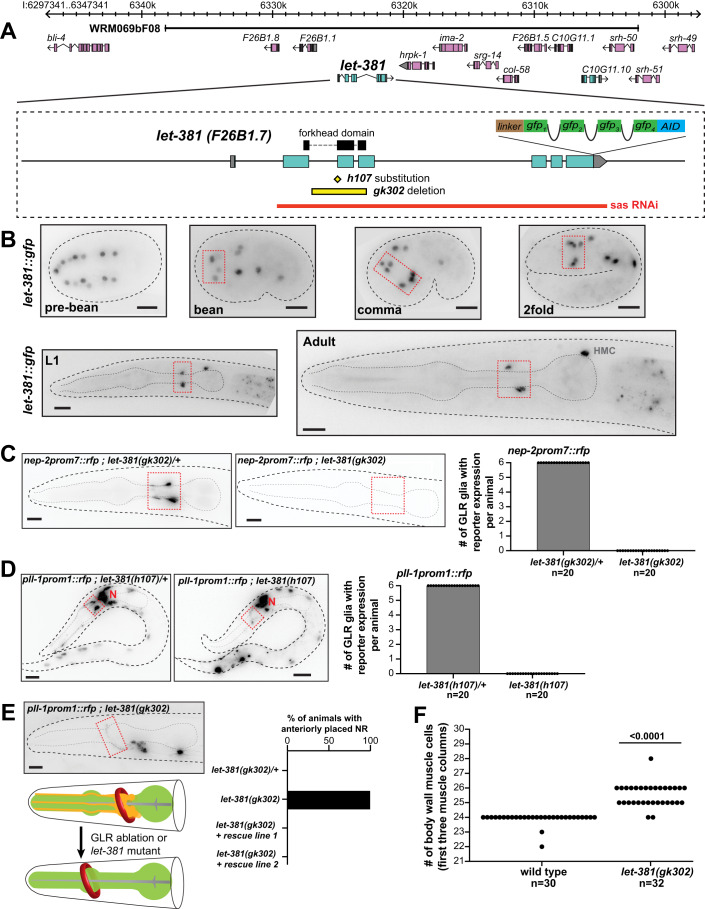
Figure 2. let-381/FoxF is required for GLR glia fate specification.
(A) let-381 genomic locus showing mutant alleles, reporters, fosmid genomic clones and RNAi sequences used in this study. (B) Expression of the endogenous let-381::gfp reporter at different stages during development. Dashed red boxes outline expression in GLR glia. (C) Absence of nep-2prom7::rfp expression in GLR glia (dashed red box) in homozygous let-381(gk302) mutants (right) as opposed to heterozygous animals (left). Quantification (number of GLR glia with nep-2prom7::rfp expression) is shown in the bar graph on the right. (D) Absence of pll-1prom1::rfp expression in GLR glia (dashed red box) in homozygous let-381(h107) mutants (right) as opposed to heterozygous animals (left). Quantification (number of GLR glia with pll-1prom1::rfp expression) is shown in the bar graph. Red “N” denotes pll-1prom1::rfp expression in neurons. (E) Similar to GLR glia-ablated animals (schematic), let-381(gk302) homozygous mutants lacking GLR glia exhibit anteriorly displaced nerve ring (NR). Dashed red box outlines a neuronal axon of the NR. Red circle in schematic indicates the Nerve Ring and pharynx is shown in green. Quantification is shown in the bar graph. Transgenic animals carrying the fosmid genomic clone WRM069bF08 with wild-type let-381 (rescue lines 1 and 2) display normal NR position. (F) Number of body wall muscle cells in the first three muscle columns of head and neck in wild type and let-381(gk302) mutants. Data information: unpaired t test was used for statistical analysis in (F). Anterior is left, dorsal is up and scale bars are 10 μm for all animal images. Source data are available online for this figure.