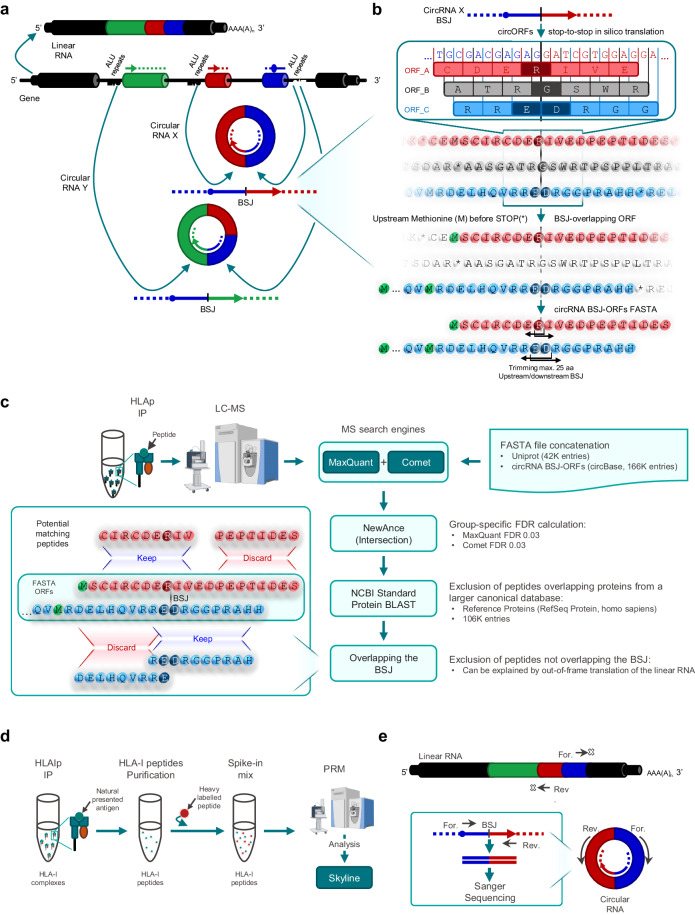
Fig. 1. Immunopeptidomics workflow for identification and validation of circRNA-derived HLA-I bound peptides.
a Canonical splicing generates a linear mRNA molecule comprising a 5’ cap and 3’ poly-A tail which assist the canonical initiation of translation. Through back-splicing, a downstream 5’ splice site is joined to an upstream 3’ splice site, producing a covalently closed structure called circRNA. In exonic circRNAs, back-splicing is associated with the presence of complementary ALU repeats in their flanking long introns15. One host gene can generate multiple circRNAs. circRNAs can undergo cap-independent translation. For illustration purposes, a circRNA composed exclusively of exons is shown and the different back-spliced exons are colored. b Fasta file construction for MS search for the identification of circRNA-derived peptides. Sequences flanking the BSJ of circRNAs were extracted from circBase and in silico translated in three forward frames. Only BSJ overlapping translated ORFs were kept, having a methionine (canonical initiation translation codon ATG) encoded upstream of the BSJ (colored in green). ORFs sequences were trimmed to a maximum of 25 amino acids upstream and downstream of the amino acid(s) encoded within the BSJ. Workflow is illustrated using a fictitious transcript sequence derived from an exonic circRNA, but the strategy was applied to all human circRNAs in circBase, regardless of their annotation. The different frames of translation are differently colored and the amino acids spanning the BSJ are highlighted with a darker color. c After immunoaffinity purification of HLA complexes, peptides were purified and analyzed by LC-MS/MS. Peptide identification was performed by two search engines against the above mentioned circRNA-derived ORFs database concatenated with a Human Uniprot database and applying a group-specific FDR. NCBI Standard Protein Blast was used to exclude peptides matching known proteins found in the RefSeq human protein database. Only peptides overlapping the BSJ were kept. d PRM validation was used to confirm the presence of the candidate peptides with heavy peptides spiked in the matrix. e At transcript level, the expression of the circRNAs was confirmed by divergent RT-PCR followed by direct Sanger Sequencing. Some elements from panels c and d were created with BioRender.com.