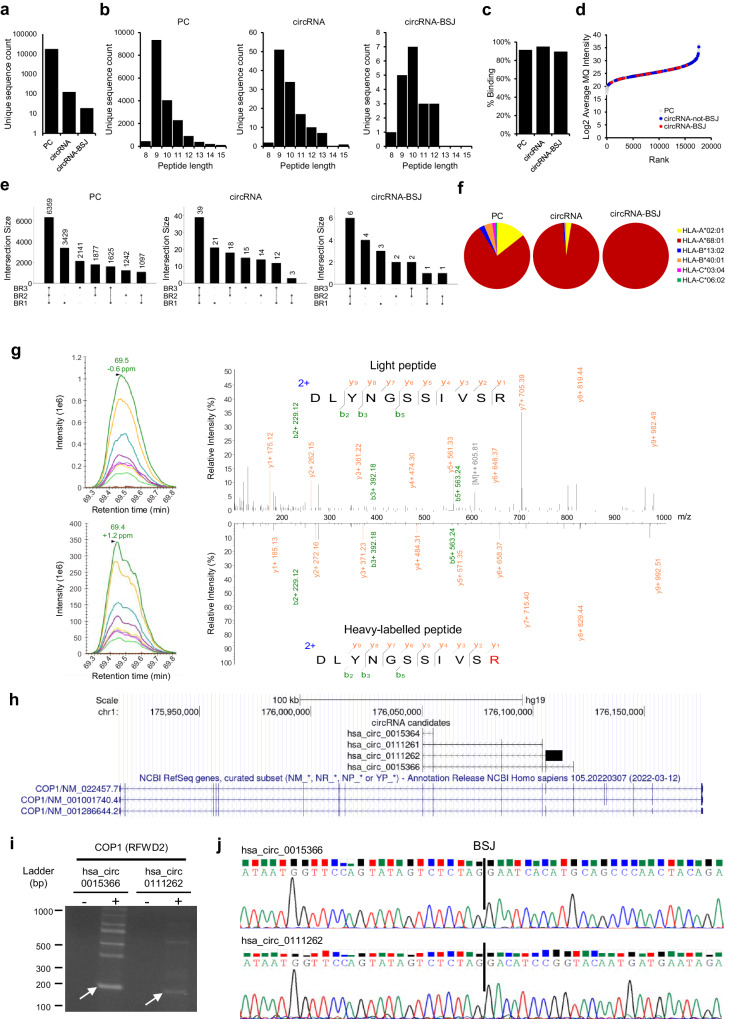
Fig. 2. Validation of circRNA-derived peptides overlapping the back-splice junction.
a Total number of peptide sequences, (b) Peptide length distribution and (c) percentage of peptides predicted as HLA binders (% rank < 2), in T1185B, annotated in the protein (PC) and circRNA groups, and circRNA-BSJ subgroup. d Rank plots of the PC, circRNA-not-BSJ, circRNA-BSJ derived peptides intensities in T1185B cell line. e UpSet plots showing the overlap of detected peptides identified in PC, and circRNA groups, and circRNA-BSJ subgroup among the three biological replicates of T1185B cell line. f HLA restriction of PC, circRNA (groups), circRNA-BSJ (subgroup) derived binder peptides in T1185B cell line. g PRM validation of peptide DLYNGSSIVS[R] encoded by circRNAs hosted by COP1 (RFWD2) gene. Skyline (left) and pLabel visualizations (right) showed the co-elution and similar MS/MS fragmentation patterns of endogenous light (above) and synthetic heavy-labelled (below) peptides. To improve readability, MS/MS fragmentation pattern figures were edited and displayed as a mirror plot. h UCSC genome browser (hg19) visualization of the four circRNAs hosted by COP1 (RFWD2) which can potentially encode peptide DLYNGSSIVS[R] within an ORF overlapping their respective BSJs. i Agarose gel electrophoresis of divergent RT-PCR products validating the expected amplicons (size) in two of the COP1 (RFWD2) circRNAs, hsa_circ_0015366 and hsa_circ_0111262. Arrows indicate the amplicons that were selected for direct Sanger Sequencing. The image was adjusted to improve legibility. This experiment was performed once. j Sanger sequencing demonstrating the specific nucleotide sequence upstream and downstream the BSJ for the above circRNAs. Source data are provided as a Source Data file.