Abstract
Background
As the most common primary bone cancer, the therapy of osteosarcoma requires further study. An anthraquinone derivative, emodin, has been found to have anticancer potential. We proposed that emodin suppresses osteosarcoma by cell cycle regulation mediated by p53.
Methods
This study determined the effect of emodin on viability and apoptosis of 6 osteosarcoma cell lines (p53 null cells MG63, G292, and A-673; p53 mutated cells HOS and SK-PN-DW; p53 expressing cells U2OS and 2 osteoblast cell lines), then knockdown p53 in U2OS, and observed the impacts of emodin on p53, p21, cyclin proteins, and cell cycle.
Results
High dose emodin (40–160 μM) induced cell death and apoptosis of all the cell lines; medium dose emodin (20 μM) preferentially inhibited osteosarcoma cells; low dose emodin (1–10 μM) preferentially inhibited p53 expressing osteosarcoma cells. Emodin dose-dependently inhibited p53 and p21 in U2OS. Emodin at 10 μM decreased the expression of Cdk2, E2F, and Cdk1; and increased RB but had no effects on cyclin E and cyclin B. The knockdown of p53 almost eliminated all the impacts of 10 μM emodin on cell cycle proteins.
Conclusions
Emodin suppresses U2OS by p53-mediated cell cycle regulation.
Keywords: p53, Cell cycle, Emodin, Osteosarcoma
1. Introduction
Cancer has been one of the most studied topics in the world [[1], [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16]], while osteosarcoma is the most pervasive primary bone cancer in the world [17]. There are about 4–6 osteosarcoma cases every one million people each year [18]. With the development of osteosarcoma therapy, the survival of osteosarcoma patients has slowly climbed over the years, yet, data suggested that more than 30% of patients can not survive in 5 years after osteosarcoma prognosis [18]. Tumor surgery is one of the major treatments for cancers [16] including osteosarcoma [19]. Optimizing the medicines used in cancer treatment has been a critical issue for clinicians [20,21]. Many efforts have been made to study the pathology and therapy of osteosarcoma [22], but, so far, the current treatments for this disease can not bring desirable therapeutic effects.
Many compounds from plants have been applied in the treatment of bone-related disease [[23], [24], [25], [26], [27]] and traditional medicine has been used for cancer treatments [6,13,28]. An anthraquinone derivative, emodin (1,3,8‐trihydroxy‐6‐methylanthraquinone), which is found as an active ingredient in several types of plants [[29], [30], [31], [32], [33]], has been found as an essential component in traditional medicines prescription for cancer treatments [34]. Pharmacological studies reveal that emodin has anti-cancer and anti-inflammatory potential [35,36]. For example, emodin can inhibit cell growth in tumor studies and result in apoptosis of MCF‐7, a commonly used breast cancer cell line. Another study showed that emodin arrested the cell cycle of SMMC-7721, a commonly used liver cancer cell line [37]. Besides, traditional herb medicines containing emodin have been used for bone disease treatments for thousands of years in China. Researchers have shown that emodin targeted bone cells and regulated bone metabolism in a mouse model of LPS‐mediated osteoporosis [38]. Hence, we suggested that emodin might potentially affect osteosarcoma.
P53, a cancer inhibitor protein, was reported to mediate the impacts of emodin on diffuse large B cell lymphoma [39]. In this study, we proposed that emodin suppresses osteosarcoma by p53-mediated cell cycle regulation. The aim of this study is to understand the role of p53 in osteosarcoma and explore the potential of emodin as a medicine for osteosarcoma.
2. Methods & materials
2.1. Tissue culture
Human osteosarcoma cell lines A-673, SK-PN-DW, MG-63, HOS, G-292, U-2 OS (U2OS), and human osteoblast cell line hFOB 1.19 were purchased from ATCC (Washington, USA). In addition, the human primary osteoblast cell line HOB was obtained from PromoCell (Heidelberg, Germany). The ATCC-formulated McCoy's 5a Medium Modified (Catalog No. 30–2007) with 10% FBS was used to culture cells. Cell lines were incubated in an incubator (5% CO2, 37 °C).
2.2. Cell viability detection
Cell viability was detected using the MTT assay, which was described previously [9]. Cells were grown on a 96-well plate at 20,000 cells/well, grown in serum-free media for 24 h, and exposed to the tested agent for 24 h. Then, methylthiazoletetrazolium (MTT, Biotium, Hayward, CA) was added (40 μL per well) for 4 h. The plate was detected with a microplate reader at 490 nm.
2.3. Drug
The emodin was purchased from Techmate Ltd (Buckinghamshire, UK). It is stored at 4 °C and dry environment before being used.
2.4. Apoptosis detection
The cell death was determined using the Cell Death Detection ELISA plus (Roche, Indianapolis, IN, USA), which is based on monitoring DNA fragmentation. Cell apoptosis level was determined by Human Bcl-2 ELISA Kit (ab119506, Abcam, Cambridge, UK). The plate was read with a microplate reader at 490 nm. Cell lines incubated at 55 °C for 20 min were used as positive controls for cell death [12]. Cells exposed to 5 μM Etoposide (ab120227, 1 mM stock prepared in DMSO, Abcam, Cambridge, UK) were used as the apoptosis-positive group.
2.5. Cell transfection
P53 protein was knocked down in U2OS cells. Briefly, cells were transfected with shRNA (h P53)-(GFP-Bsd) lentivirus (LVP343-GB, GenTarget, San Diego, CA, USA), or negative shRNA (SIC001, Sigma-Aldrich, MO, USA) using Lipofectamine® 2000, as described elsewhere [40,41]. The transfecting medium was replaced by a normal medium after 8 h. The expression of p53 in cells was validated by immunofluorescence experiment and western blotting assay 48 h after the transfection.
2.6. Cell cycle analysis
The propidium iodide (PI) staining was used to determine the cell cycle with flow cytometry [42]. Cold PBS was used to wash cells. Then, cells were fixed in 70% ethanol for 1.5 min at 4 °C. Cold PBS was used to wash cells again. Then, cells were stained with propidium iodide at 37 °C for 30 min (0.02 mg/mL in cold PBS with 0.2 mg/mL DNAse-free RNAse A and 0.1% Triton X-100). Stained cells were analyzed on the BD FACSCalibur (Becton Dickinson. San Jose, CA, USA).
2.7. Western blotting
Protein was detected by western blotting experiments [43]. Cells were collected and the protein in samples was extracted using RIPA plus protease inhibitor (Sigma-Aldrich, MO, USA). The protein concentration was determined using the BCA assay kit(ab102536, Abcam, Cambridge, UK). The separation of proteins with different sizes was achieved by SDS gel electrophoresis with a loading amount of 20 μg/well. The proteins were then transferred onto membranes for blotting experiments. The membranes were incubated in 5% skimmed milk in TBST (Sigma-Aldrich, MO, USA). Then, the membranes were then incubated in primary antibodies (1:1000 dilution of BSA, 4 °C, overnight; Anti-p53 antibody ab26, Anti-p21 antibody ab109520, Anti-Cdk2 antibody ab32147, Anti-Cyclin E antibody ab33911, Anti-Rb antibody ab32513, Anti-E2F1 antibody ab4070, Anti-CDK1 antibody ab133327, Anti-Cyclin B1 antibody ab181593), and secondary antibodies (Goat Anti-Rabbit HRP ab205718, Rabbit Anti-Mouse HRP ab6728, 1:3000 dilution of BSA, room temperature, 2 h). All antibodies were obtained from Abcam (Cambridge, UK). Enhanced chemiluminescent detection reagents (Sigma-Aldrich, MO, USA) were used for the visualization of the proteins on the membrane.
2.8. Immunofluorescence
Immunofluorescence was used to observe the level of p53 in cells [44]. Cells were fixed and permeabilized in 4% formaldehyde and 0.1% Triton X-100 subsequently. Cells were incubated first with an anti-p53 antibody (ab32389, Abcam, Cambridge, UK) and then with an EGFP-labeled secondary antibody (Goat Anti-Rabbit Alexa Fluor® 488 ab150077). An inverted fluorescence microscope (Leica, Germany) was used to observe the cells.
2.9. Statistics and plotting
The difference in the results was analyzed using a T-test or one-way ANOVA and Dunnett's post hoc tests: p < 0.01 indicates a significant difference. Means and stander deviation were plotted using GraphPad Prism software.
3. Results
3.1. Emodin preferentially suppressed the viability of p53-expressing osteosarcoma cells
Emodin at 20–80 μM was previously adopted in a liver tumor project [37]. In this study, we designed experiments to test the effect of emodin concentration at a larger concentration range, from 1 to 160 μM, on 6 osteosarcoma cell lines, HOS, MG-63, G-292, A-673, SK-PN-DW, and U2OS, and 2 osteoblast cell lines hFOB and HOB. MTT assay is commonly applied in cancer research [45]. Cells were grown on a 96-well plate at 20,000 cells/well when conducting treatment. Data showed that, after 24 h of exposure, emodin at 40–160 μM significantly reduced the viability of all the cell lines. Emodin at 40 μM preferentially inhibited osteosarcoma cells (inhibited over 50% of viability) over normal osteoblast cells (inhibited about 30% of viability). Emodin at 20 μM significantly suppressed the viability of HOS, SK-PN-DW, and U2OS. U2OS was the most sensitive cell line. Its effective concentration was 1 μM (Fig. 1). MG63 (p53 null) [46], G292 (p53 null) [47], A-673(p53 null) [48] are p53 muted, HOS (p53-R156P) [49] and SK-PN-DW(p53-C176F) [50] are p53 mutated, while U2OS (wild type p53) [51] have wildtype p53. Results showed that emodin at 1–10 μM preferentially suppressed p53 expressing osteosarcoma cell lines more than p53 muted or mutated osteosarcoma cells. As we were interested in p53 in emodin actions, U2OS was used in the subsequent study because it expressed p53 and it was the most sensitive cell line for emodin viability inhibition based on our results (Fig. 1A–H). These results indicated that emodin at a high concentration had an inhibition effect on all cells, but at a lower concentration, it preferentially suppressed p53 expressing osteosarcoma cell lines than other cells.
Fig. 1.
Effect of emodin on the cell viability of osteosarcoma and osteoblast cell lines. Cells were exposed to emodin for 24 h and the viability was determined using an MTT assay. “*” indicates a significant difference (p < 0.01) compared with 0 μM emodin. A-F. osteosarcoma cell lines. G-H. osteoblast cell lines.
3.2. Emodin-induced cell death and apoptosis of all cells at a high concentration
To test whether emodin at high concentration has toxicity toward cells, we determined the cell death after 24 h of exposure to 1–160 μM emodin. Cell lines incubated at 55 °C for 20 min which were regarded as 100% dead cells were used as positive controls. Cells were grown on a 96-well plate at 20,000 cells/well when conducting treatment. Results revealed a significant cell death induction of emodin at 40–160 μM among all the cell lines, while at doses lower than 20 μM, emodin showed no significant difference (Fig. 2A–H). Thus, the viability inhibition effect of emodin at high concentrations on cells resulted from cell death. To further identify if the cell death resulted from apoptosis, we determined the Bcl-2 level in cell lines. Data suggested that emodin at 40–160 μM significantly decreased the level of Bcl-2 among all the cell lines, while emodin at 0–20 μM failed to generate significant differences (Fig. 3A–H). These results indicated that the cell death caused by the high concentration of emodin was a result of the apoptosis alteration, but emodin at 0–20 μM did not induce cell death or apoptosis in all cell lines. Hence, in the subsequent study, emodin at 10 μM was used.
Fig. 2.
Effect of emodin on cell death of osteosarcoma and osteoblast cell lines. Cells were exposed to emodin for 24 h and the cell death was determined using Cell Death Detection ELISA. Cell lines incubated at 55 °C for 20 were used as positive controls (PC) for cell death. “*” indicates a significant difference (p < 0.01) compared with 0 μM emodin. A-F. osteosarcoma cell lines. G-H. osteoblast cell lines.
Fig. 3.
Effect of emodin on apoptosis of osteosarcoma and osteoblast cell lines. Cells were treated for 24 h and the apoptosis was determined by Bcl-2 ELISA. Cell exposed to 5 μM Etoposide for 12 h was used as a positive control (PC) for apoptosis. “*” indicates a significant difference (p < 0.01) compared with 0 μM emodin. A-F. osteosarcoma cell lines. G-H. osteoblast cell lines.
3.3. Emodin inhibited p53 and p21 proteins
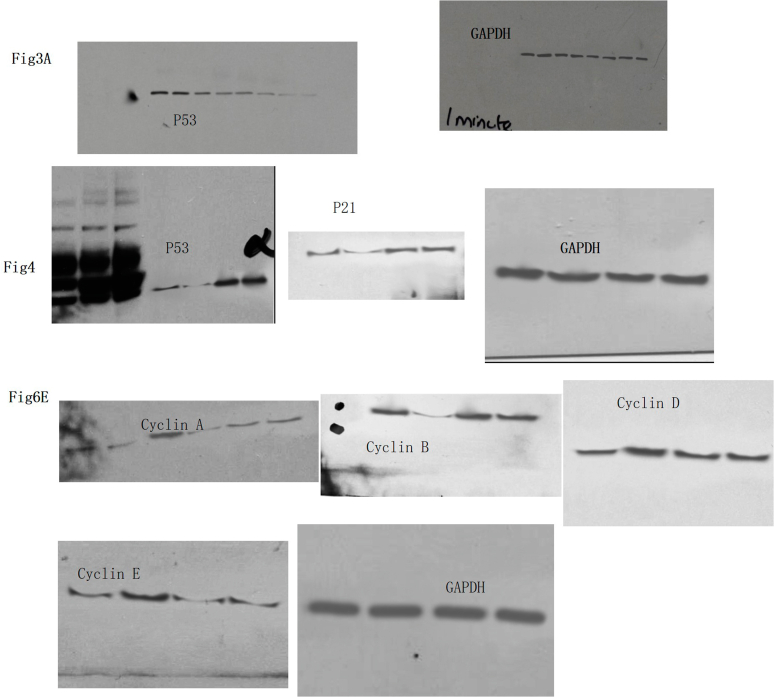
P53 impacted the inhibition of emodin toward diffuse large B cell lymphoma [39]. We first determined the impacts of emodin on p53 and p21 levels in a p53-expressing osteosarcoma cell line U2OS. Results showed that emodin dose-dependently inhibited p53 and p21 levels in U2OS cells (Fig. 4ABC). We stained U2OS cells with an anti-p53 antibody and confirmed that emodin decreased p53 protein expression (Fig. 4 D left panel). We believed that the effect of emodin on U2OS cells was mediated by p53/p21. Then we knocked down p53 in U2OS to investigate the impacts of emodin. After the transfection, p53 was decreased by about 90% (Fig. 4 DEF). The p53 level in transfected cells was not significantly decreased by emodin, indicating that p53 knockdown offset the effect of emodin. Therefore, p53 is a potential key regulator in the impacts of emodin on U2OS cells.
Fig. 4.
Effect of emodin on p53 and p21 in U2OS. U2OS cells were exposed to emodin at different concentrations and then the level of p53 and p21 were determined using western blotting. A. representative image of western blotting. B. Effect of emodin on p53 in U2OS. C. Effect of emodin on p21 in U2OS. D. Immunofluorescence of p53 knockdown U2OS. The emodin treatment group was treated with emodin for 24 h. E.F. Effect of emodin on p53 expression of p53 knocked down U2OS cells. “*” indicates a significant difference (p < 0.01). Non-adjusted images were provided as supplementary materials.
3.4. Emodin affected the viability of U2OS through the cell cycle
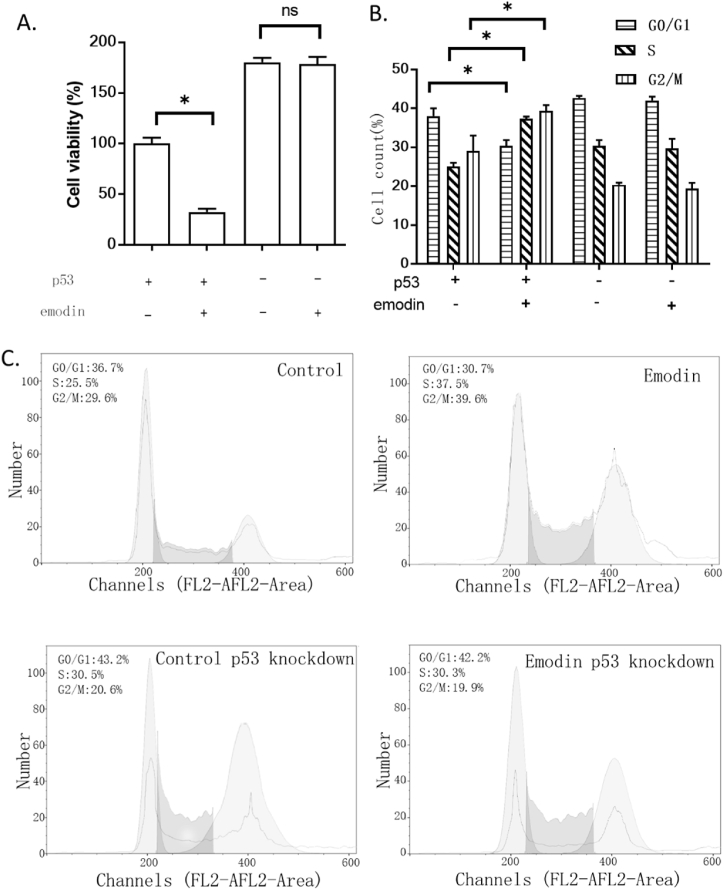
To study the impacts of p53 on emodin pharmacological actions, this study determined the impacts of emodin on p53 knockdown U2OS cells. Results showed that p53 knockdown almost locked the effect of emodin (Fig. 5A). We proposed that the effect of emodin was mediating the cell cycle, hence, we observed alterations of the cell cycle in U2OS cells. Results showed that emodin decreased cell number in the G1/G0 phase and increased cell number in the S and G2/M phases. The G0/G1 phase is the phase for cells to process other activities besides proliferation such as differentiation [52], while the S/G2/M phase is the phase for cells to proliferate [53]. These observations further confirmed that emodin had an impact on cell growth. In addition, the knockdown of p53 eliminated the sensitivity of U2OS toward emodin, revealing that p53 might be involved in cell cycle regulation (Fig. 5BCE). This conclusion agreed with a previous study suggesting that p53/p21 impacted genes regulating the G2/M phases [54].
Fig. 5.
The effect of emodin on the viability and cell cycle of the U2OS. A. The effect of emodin on the viability of p53 knockdown U2OS cells. The cell viability was determined using an MTT assay. B. the effect of emodin on the cell cycle of p53 knockdown U2OS cells. The cell cycle was analyzed by flow cytometry with PI staining. “*” indicates a significant difference (p < 0.01). C. Representative images of cell cycle analysis.
3.5. Emodin affected cell cycle proteins
To confirm the role of p53, we also detected some regulators of the cell cycle, including p21, Cdk2, cyclin E, RB, E2F, Cdk1, and cyclin B. Emodin reduced the expression of Cdk2, E2F, and Cdk1. Emodin also increased RB but had no significant effects on cyclin E and cyclin B. The knockdown of p53 almost eliminated all the effects of emodin (Fig. 6). This result accounted for the previous cell cycle alternation, indicating that p53 might mediate the impacts of emodin on the cell cycle (Fig. 6A–H).
Fig. 6.
Effect of emodin on cell cycle regulators in U2OS. A. Representative images of western blotting. B–H. Western blotting results of p21, Cdk2, cyclin E, RB, E2F, Cdk1, and cyclin B. “*” indicates a significant difference (p < 0.01). Non-adjusted images were provided as supplementary materials.
4. Discussion
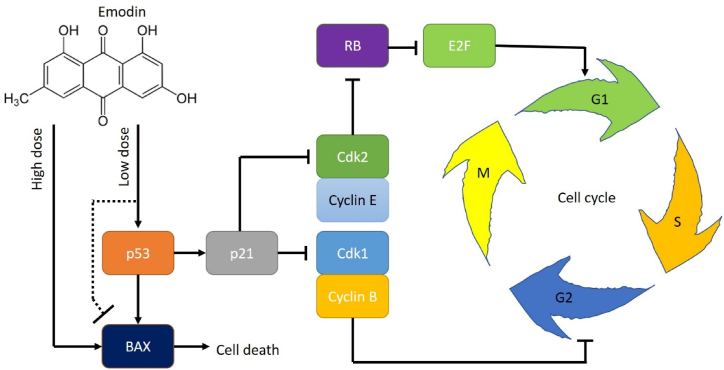
Cancer is one of the top popular topics in medical studies. Many studies apply bioinformatics to investigate cancer [1,28,[55], [56], [57], [58], [59]] as well as other diseases [60,61], yet, experimental evidence is required for validation of these mechanisms [4]. In this study, we found the impacts of emodin on the proliferation of U2OS. As we mentioned, this is a p53-expressing osteosarcoma cell line. Cell viability was detected using the widely used MTT assay [45]. By comparing its effect on osteosarcoma and osteoblast cells, we obtained the effective concentration range: emodin at a concentration over 40 μM induced cell death in both osteosarcoma and osteoblast cells; concentration at 20 μM only suppressed osteosarcoma cells; concentration at 1–10 μM preferentially inhibit the viability of p53 expressing cells. The sensitivity of U2OS toward emodin is higher than that of liver cancer cell line SMMC-7721 [37]. Cell death assay confirmed that emodin at a concentration higher than 40 μM significantly induced cell death and apoptosis. The induction of apoptosis was not distinguished between osteosarcoma and osteoblast or p53 muted/mutated osteosarcoma and p53 expressing osteosarcoma. This suggested that high-dose emodin impacts cells by inducing cell death and apoptosis through other mechanisms besides p53 or other cancer-specific targets. Yet, emodin at 1–10 μM only suppressed the viability of p53 expressing osteosarcoma cells suggesting that low-dose emodin impacted osteosarcoma cells via p53. Interestingly, p53 was supposed to have regulatory effects on apoptosis [62], but our result showed that low-dose emodin affected p53 without altering apoptosis, thus we suggested that low-dose emodin might have an inhibition effect on apoptosis BAX protein (Fig. 7).
Fig. 7.
Proposed regulatory mechanism of emodin in bone cells.
A previous study had shown that p53 plays a role in the inhibition of emodin toward diffuse large B cell lymphoma [39]. P53 was proved to be a key regulator for the cell cycle in osteosarcomas [63]. We tested this in U2OS. Emodin caused a significant alteration in the cell cycle of U2OS. Both G0/G1 and s/G2/M were altered by emodin indicating that emodin suppressed osteosarcoma cell proliferation by regulating the cell cycle.
In cancer, the p53/p21 pathway negatively regulates cdk1 and cyclin B, which regulates the processes of the G2 phase [64]. The increase of p53 upregulates the p21, which affects the function of the cyclin B/cdk1 complex [65]. In this study, the knockdown of p53 eliminated the effects of emodin on osteosarcoma cells, revealing that p53 affected the cell cycle regulation by emodin. This conclusion was in line with a previous study that showed that p53/p21 regulated the G2/M cell cycle regulator genes [54]. In this study, emodin significantly decreased p53 expression. We suggested that the decrease of p53 by emodin further decreased p21 expression and down-regulated cells in the G2 phase. Besides, the cyclin E/CDK2 complex might be inactivated by p21 [66]. E/CDK2 complex is responsible for regulating cells from the G1 phase entering the S phase, which might account for the change in the G1 phase and the S phase. In this study, cyclin E was not affected by emodin but as the Cdk2 was decreased, it resulted in less inhibition of RB and further caused a decrease in E2F protein, which was regulated by RB [67]. We tested the major E2F protein E2F1 because it is one of the most cancer-relative proteins in the E2F family [68]. Thus, the decreased E2F inhibited the G1 checkpoint [69]. On the other hand, p21 also inhibited Cdk1 and this led to a decrease of the Cdk1/cyclin B complex, even though the level of cyclin B was not affected. The inhibition of the Cdk1/cyclin B complex can facilitate the G2 activities [[70], [71], [72]] (Fig. 7).
However, in this study, not all the osteosarcoma cell lines we tested expressed p53: MG63 (p53 null) [46], G292 (p53 null) [47], A-673(p53 null) [48] are p53 muted, HOS (p53-R156P) [49] and SK-PN-DW(p53-C176F) [50] are p53 mutated, while U2OS (wild type p53) [51] have wildtype p53. Our results showed that p53 mutated cells and p53 muted cells reacted to emodin differently. We suggested that p53 expressing osteosarcoma had higher sensitivity toward emodin, which might account for different clinical therapeutic effects of emodin on osteosarcoma patients. While different cell lines might have different mechanisms, this study only indicated that p53-mediated cell cycle regulation is a mechanism of emodin in U2OS. However, the other cell lines in this study might have alternative mechanisms that have not been identified in this study, especially for the cell death induction of emodin at high doses. We suggested that p53 is not an essential regulator in the effect of emodin, but it is critical for the sensitivity of osteosarcoma toward emodin. Besides, whether p53 is a direct or indirect target of emodin is still unclear and requires more exploration. P53 has been found to be involved in cancer stem cells [73] and ion channel regulation [74,75], thus, we suggested that cancer stem cells [76] and ion channels [14,77] might be the mechanisms underlying these emodin effects. Further study is required to test if this mechanism is common among different cancer types.
While this study provides valuable insights into the potential anti-osteosarcoma effects of emodin and its interaction with p53-mediated cell cycle regulation, there are certain limitations that should be acknowledged: The study primarily focused on the U2OS cell line, which expresses wild-type p53. Different osteosarcoma cell lines with varying p53 statuses demonstrated distinct responses to emodin. The findings may not fully represent the heterogeneity of osteosarcoma, and further investigations with a broader spectrum of cell lines could enhance the generalizability of the results. The current study relies on in vitro experiments, and the effects observed may differ in the complex in vivo environment. Future studies incorporating animal models or clinical trials are essential to validate the efficacy and safety of emodin as a potential therapeutic agent for osteosarcoma. The mechanisms underlying the effects of emodin on osteosarcoma cells are likely multifaceted. This study primarily focused on p53-mediated cell cycle regulation, but additional pathways and molecular targets may contribute to emodin's anti-cancer properties. Further exploration of these potential mechanisms is warranted. Translating findings from laboratory studies to clinical applications presents challenges. The pharmacokinetics, bioavailability, and potential side effects of emodin in human subjects need thorough investigation before considering it as a mainstream therapeutic option for osteosarcoma. The study identified an effective concentration range for emodin, but the optimal dosage for therapeutic purposes remains uncertain. Determining a dose that maximizes anti-cancer effects while minimizing toxicity is crucial for future clinical applications.
For future prospects, addressing the limitations of the current study and paving the way for the development of emodin as a viable treatment option for osteosarcoma requires a multifaceted approach. Future research should delve into alternative molecular mechanisms and signaling pathways through which emodin impacts osteosarcoma cells. This exploration could encompass investigating its effects on cancer stem cells, ion channels, and other relevant targets to gain a more comprehensive understanding of the underlying mechanisms. Moreover, the progression from in vitro experiments to clinical applications necessitates rigorous preclinical studies using animal models. Evaluating the efficacy, safety, and pharmacokinetics of emodin in a more complex in vivo environment is crucial before considering its potential in human subjects. These preclinical investigations will provide essential insights into the translational potential of emodin. Taking the next step, clinical trials should be initiated to systematically assess the feasibility, safety, and efficacy of emodin as either an adjunctive or standalone therapy for osteosarcoma patients. These trials will offer valuable data on the real-world applicability and potential benefits of emodin in a clinical setting. Additionally, exploring combination therapies is essential to unlock potential synergies between emodin and existing osteosarcoma treatments. This approach aims to enhance therapeutic outcomes and mitigate the development of resistance, addressing a critical challenge in cancer treatment. Concurrently, conducting dose-response studies will be pivotal in identifying the optimal concentration of emodin for anti-osteosarcoma effects while minimizing adverse effects. This dose optimization is a crucial step in establishing the therapeutic window and ensuring the safety and efficacy of emodin in a clinical context. By addressing these aspects comprehensively, future research endeavors can contribute significantly to unraveling emodin's therapeutic potential for osteosarcoma, potentially paving the way for its development into a clinically viable and effective treatment option.
Funding
None.
Additional information
No additional information is available for this paper.
CRediT authorship contribution statement
Qian Zhang: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Shuli Hao: Writing – review & editing, Writing – original draft, Visualization, Supervision, Methodology, Formal analysis, Data curation. Guangyou Wei: Writing – review & editing, Writing – original draft, Validation, Resources, Formal analysis. Xiangyu Liu: Writing – original draft, Visualization, Validation, Investigation. Yang Miao: Writing – original draft, Supervision, Formal analysis, Data curation.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e26850.
Contributor Information
Qian Zhang, Email: QZhoncology@gmail.com.
Yang Miao, Email: yangmiao004@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
References
- 1.Liu H., Weng J. A pan-cancer bioinformatic analysis of RAD51 regarding the values for diagnosis, prognosis, and therapeutic prediction. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.858756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H., Weng J. Gene; 2022. A Comprehensive Bioinformatic Analysis of Cyclin-dependent Kinase 2 (CDK2) in Glioma. [DOI] [PubMed] [Google Scholar]
- 3.Liu H., Tang T. Pan-cancer genetic analysis of cuproptosis and copper metabolism-related gene set. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.952290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H., Dilger J.P., Lin J. Pharmacology & Therapeutics; 2022. A Pan-Cancer-Bioinformatic-Based Literature Review of TRPM7 in Cancers. [DOI] [PubMed] [Google Scholar]
- 5.Liu H. Pan-cancer profiles of the cuproptosis gene set. Am. J. Cancer Res. 2022;12(8):4074–4081. [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H. Toxic medicine used in Traditional Chinese Medicine for cancer treatment: are ion channels involved? J. Tradit. Chin. Med. 2022;42(6):1019–1022. doi: 10.19852/j.cnki.jtcm.20220815.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Liu H. Cancer Biomark; 2022. Clinical Powers of Aminoacyl tRNA Synthetase Complex Interacting Multifunctional Protein 1 (AIMP1) for Head-Neck Squamous Cell Carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Z., et al. Recent Pat Anticancer Drug Discov; 2022. Potential Therapeutic Application of Local Anesthetics in Cancer Treatment. [DOI] [PubMed] [Google Scholar]
- 9.Liu H., Dilger J.P., Lin J. Lidocaine suppresses viability and migration of human breast cancer cells: TRPM7 as A target for some breast cancer cell lines. Cancers. 2021;13(2):234. doi: 10.3390/cancers13020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Liu H., Han Y. Research Square; 2021. Potential Roles of Cornichon Family AMPA Receptor Auxiliary Protein 4 (CNIH4) in Head and Neck Squamous Cell Carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H., Dilger J.P., Lin J. Effects of local anesthetics on cancer cells. Pharmacol. Ther. 2020;212 doi: 10.1016/j.pharmthera.2020.107558. [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Dilger J.P., Lin J. The role of transient receptor potential melastatin 7 (TRPM7) in cell viability: a potential target to suppress breast cancer cell cycle. Cancers. 2020;12(1) doi: 10.3390/cancers12010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H. Effect of traditional medicine on clinical cancer. Biomedical Journal of Scientific & Technical Research. 2020;30(4):23548–23551. [Google Scholar]
- 14.Liu H. Nav channels in cancers: nonclassical roles. Global Journal of Cancer Therapy. 2020;6(1):5. [Google Scholar]
- 15.Liu H. A prospective for the potential effect of local anesthetics on stem-like cells in colon cancer. Biomedical Journal of Scientific & Technical Research. 2020;25(2):18927–18930. [Google Scholar]
- 16.Liu H. A clinical mini-review: clinical use of Local anesthetics in cancer surgeries. The Gazette of Medical Sciences. 2020;1(3):30–34. [Google Scholar]
- 17.Ottaviani G., Jaffe N. Pediatric and Adolescent Osteosarcoma. Springer; 2009. The epidemiology of osteosarcoma; pp. 3–13. [Google Scholar]
- 18.Ottaviani G., Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 19.Anderson M.E. Update on survival in osteosarcoma. Orthop. Clin. N. Am. 2016;47(1):283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 20.Li R., et al. Effect of Propofol on breast Cancer cell, the immune system, and patient outcome. BMC Anesthesiol. 2018;18(1):77. doi: 10.1186/s12871-018-0543-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R., et al. Comparing volatile and intravenous anesthetics in a mouse model of breast cancer metastasis. Proceedings of the American Association for Cancer Research Annual Meeting 2018. 2018:2162. American Association for Cancer Research. [Google Scholar]
- 22.Prater S., McKeon B. StatPearls. StatPearls Publishing Copyright © 2020, StatPearls Publishing LLC; Treasure Island (FL: 2020. Cancer, osteosarcoma. [Google Scholar]
- 23.Chen G., et al. Antiosteoporotic effect of icariin in ovariectomized rats is mediated via the Wnt/beta-catenin pathway. Exp. Ther. Med. 2016;12(1):279–287. doi: 10.3892/etm.2016.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., et al. Effects of water extract from epimedium on neuropeptide signaling in an ovariectomized osteoporosis rat model. J. Ethnopharmacol. 2018;221:126–136. doi: 10.1016/j.jep.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 25.Liu H., et al. Icariin improves osteoporosis, inhibits the expression of PPARgamma, C/EBPalpha, FABP4 mRNA, N1ICD and jagged1 proteins, and increases Notch2 mRNA in ovariectomized rats. Exp. Ther. Med. 2017;13(4):1360–1368. doi: 10.3892/etm.2017.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haixia W., et al. Effectiveness associated with different therapies for senile osteopo-rosis: a network Meta-analysis. J. Tradit. Chin. Med. 2020;40(1):17–27. [PubMed] [Google Scholar]
- 27.Wang C., et al. Effect of herba epimedium extract on bone mineral density and microstructure in ovariectomised rat. J. Pharmaceut. Biomed. Sci. 2016;6(5) [Google Scholar]
- 28.Hengrui L. An example of toxic medicine used in Traditional Chinese Medicine for cancer treatment. J. Tradit. Chin. Med. 2023;43(2):209–210. doi: 10.19852/j.cnki.jtcm.2023.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y.-C., Lim M.-Y., Lee H.-S. Emodin isolated from Cassia obtusifolia (Leguminosae) seed shows larvicidal activity against three mosquito species. J. Agric. Food Chem. 2003;51(26):7629–7631. doi: 10.1021/jf034727t. [DOI] [PubMed] [Google Scholar]
- 30.Naqvi S., Ullah M., Hadi S. 2010. DNA Degradation by Aqueous Extract of Aloe Vera in the Presence of Copper Ions. [PubMed] [Google Scholar]
- 31.Lee M.-H., Kao L., Lin C.-C. Comparison of the antioxidant and transmembrane permeative activities of the different Polygonum cuspidatum extracts in phospholipid-based microemulsions. J. Agric. Food Chem. 2011;59(17):9135–9141. doi: 10.1021/jf201577f. [DOI] [PubMed] [Google Scholar]
- 32.Wang J.-b., et al. Hepatotoxicity or hepatoprotection? Pattern recognition for the paradoxical effect of the Chinese herb Rheum palmatum L. in treating rat liver injury. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M., et al. Lipid regulation effects of Polygoni Multiflori Radix, its processed products and its major substances on steatosis human liver cell line L02. J. Ethnopharmacol. 2012;139(1):287–293. doi: 10.1016/j.jep.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 34.Hsu S.C., Chung J.G. Anticancer potential of emodin. Biomedicine. 2012;2(3):108–116. doi: 10.1016/j.biomed.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrimali D., et al. Targeted abrogation of diverse signal transduction cascades by emodin for the treatment of inflammatory disorders and cancer. Cancer Lett. 2013;341(2):139–149. doi: 10.1016/j.canlet.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 36.Wei W.-T., et al. The distinct mechanisms of the antitumor activity of emodin in different types of cancer. Oncol. Rep. 2013;30(6):2555–2562. doi: 10.3892/or.2013.2741. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X., et al. Inhibitory effect of emodin on human hepatoma cell line SMMC-7721 and its mechanism. Afr. Health Sci. 2015;15(1):97–100. doi: 10.4314/ahs.v15i1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang D.M., et al. CT imaging biomarker for evaluation of emodin as a potential drug on LPS-mediated osteoporosis mice. Acad. Radiol. 2014;21(4):457–462. doi: 10.1016/j.acra.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y., et al. Integration of bioinformatics and experiments to identify TP53 as a potential target in Emodin inhibiting diffuse large B cell lymphoma. Biomed. Pharmacother. 2018;107:226–233. doi: 10.1016/j.biopha.2018.07.168. [DOI] [PubMed] [Google Scholar]
- 40.Clements B.A., et al. A comparative evaluation of poly-l-lysine-palmitic acid and Lipofectamine™ 2000 for plasmid delivery to bone marrow stromal cells. Biomaterials. 2007;28(31):4693–4704. doi: 10.1016/j.biomaterials.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 41.Li X., et al. Changes in related circular RNAs following ERbeta knockdown and the relationship to rBMSC osteogenesis. Biochem. Biophys. Res. Commun. 2017;493(1):100–107. doi: 10.1016/j.bbrc.2017.09.068. [DOI] [PubMed] [Google Scholar]
- 42.Li R., et al. Effects of local anesthetics on breast cancer cell viability and migration. BMC Cancer. 2018;18(1):666. doi: 10.1186/s12885-018-4576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X., et al. Postmenopausal osteoporosis is associated with the regulation of SP, CGRP, VIP, and NPY. Biomed. Pharmacother. 2018;104:742–750. doi: 10.1016/j.biopha.2018.04.044. [DOI] [PubMed] [Google Scholar]
- 44.Wu Z., et al. Icaritin induces MC3T3-E1 subclone14 cell differentiation through estrogen receptor-mediated ERK1/2 and p38 signaling activation. Biomed. Pharmacother. 2017;94:1–9. doi: 10.1016/j.biopha.2017.07.071. [DOI] [PubMed] [Google Scholar]
- 45.Hengrui Liu J.P.D., Lin Jun. Pharmacology & Therapeutics; 2020. Effects of Local Anesthetics on Cancer Cells. in press. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y., et al. Arsenic trioxide induces apoptosis of p53 null osteosarcoma MG63 cells through the inhibition of catalase. Med. Oncol. 2012;29(2):1328–1334. doi: 10.1007/s12032-011-9848-5. [DOI] [PubMed] [Google Scholar]
- 47.Chandar N., et al. Inactivation of p53 gene in human and murine osteosarcoma cells. Br. J. Cancer. 1992;65(2):208–214. doi: 10.1038/bjc.1992.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aryee D.N., et al. Variability in functional p53 reactivation by PRIMA-1(Met)/APR-246 in Ewing sarcoma. Br. J. Cancer. 2013;109(10):2696–2704. doi: 10.1038/bjc.2013.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganjavi H., et al. Adenovirus-mediated p53 gene therapy in osteosarcoma cell lines: sensitization to cisplatin and doxorubicin. Cancer Gene Ther. 2006;13(4):415–419. doi: 10.1038/sj.cgt.7700909. [DOI] [PubMed] [Google Scholar]
- 50.Yu X., et al. Small molecule restoration of wildtype structure and function of mutant p53 using a novel zinc-metallochaperone based mechanism. Oncotarget. 2014;5(19):8879–8892. doi: 10.18632/oncotarget.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allan L.A., Fried M. p53-dependent apoptosis or growth arrest induced by different forms of radiation in U2OS cells: p21WAF1/CIP1 repression in UV induced apoptosis. Oncogene. 1999;18(39):5403–5412. doi: 10.1038/sj.onc.1202931. [DOI] [PubMed] [Google Scholar]
- 52.Dalton S., Coverdell P.D. Linking the cell cycle to cell fate decisions. Trends Cell Biol. 2015;25(10):592–600. doi: 10.1016/j.tcb.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., et al. Ochratoxin A-induced apoptosis of IPEC-J2 cells through ROS-mediated mitochondrial permeability transition pore opening pathway. J. Agric. Food Chem. 2017;65(48) doi: 10.1021/acs.jafc.7b04434. acs.jafc.7b04434. [DOI] [PubMed] [Google Scholar]
- 54.Fischer M., et al. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016;44(1):164–174. doi: 10.1093/nar/gkv927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H., Tang T. MAPK signaling pathway-based glioma subtypes, machine-learning risk model, and key hub proteins identification. Sci. Rep. 2023;13(1) doi: 10.1038/s41598-023-45774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H., Tang T. Pan-cancer genetic analysis of disulfidptosis-related gene set. Cancer Genet. 2023;278–279:91–103. doi: 10.1016/j.cancergen.2023.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Liu H., Tang T. A bioinformatic study of IGFBPs in glioma regarding their diagnostic, prognostic, and therapeutic prediction value. Am J Transl Res. 2023;15(3):2140–2155. [PMC free article] [PubMed] [Google Scholar]
- 58.Liu H., Tang T. bioRxiv; 2023. Pan-cancer Genetic Analysis of Disulfidptosis-Related Gene Set; p. 2023. 02. 25.529997. [DOI] [PubMed] [Google Scholar]
- 59.Liu H. Expression and potential immune involvement of cuproptosis in kidney renal clear cell carcinoma. Cancer Genetics. 2023;274–275:21–25. doi: 10.1016/j.cancergen.2023.03.002. [DOI] [PubMed] [Google Scholar]
- 60.Liu H., et al. Exploring the mechanism underlying hyperuricemia using comprehensive research on multi-omics. Sci. Rep. 2023;13(1):7161. doi: 10.1038/s41598-023-34426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H. Association between sleep duration and depression: a Mendelian randomization analysis. J. Affect. Disord. 2023;335:152–154. doi: 10.1016/j.jad.2023.05.020. [DOI] [PubMed] [Google Scholar]
- 62.Chipuk J.E., et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303(5660):1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 63.Diller L., et al. p53 functions as a cell cycle control protein in osteosarcomas. Mol. Cell Biol. 1990;10(11):5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma, Y., et al., Biological functions and clinical significance of the newly identified long non-coding RNA RP1-85F18.6 in colorectal cancer. Oncol. Rep.. [DOI] [PMC free article] [PubMed]
- 65.Marine J.C., Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17(1):93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 66.Minella A.C., et al. p53 and p21 form an inducible barrier that protects cells against cyclin E-cdk2 deregulation. Curr. Biol. 2002;12(21):1817–1827. doi: 10.1016/s0960-9822(02)01225-3. [DOI] [PubMed] [Google Scholar]
- 67.Engeland K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022;29(5):946–960. doi: 10.1038/s41418-022-00988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang Z., et al. ERINA is an estrogen-responsive LncRNA that drives breast cancer through the E2F1/RB1 pathway. Cancer Res. 2020;80(20):4399–4413. doi: 10.1158/0008-5472.CAN-20-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsumura I., Tanaka H., Kanakura Y. E2F1 and c-Myc in cell growth and death. Cell Cycle. 2003;2(4):333–338. [PubMed] [Google Scholar]
- 70.Timofeev O., et al. Cdc25 phosphatases are required for timely assembly of CDK1-cyclin B at the G2/M transition. J. Biol. Chem. 2010;285(22):16978–16990. doi: 10.1074/jbc.M109.096552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Y., et al. c-Myc regulates the CDK1/cyclin B1 dependent-G2/M cell cycle progression by histone H4 acetylation in Raji cells. Int. J. Mol. Med. 2018;41(6):3366–3378. doi: 10.3892/ijmm.2018.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Catania M.G., Mischel P.S., Vinters H.V. Hamartin and tuberin interaction with the G2/M cyclin-dependent kinase CDK1 and its regulatory cyclins A and B. J. Neuropathol. Exp. Neurol. 2001;60(7):711–723. doi: 10.1093/jnen/60.7.711. [DOI] [PubMed] [Google Scholar]
- 73.Velletri T., et al. P53 functional abnormality in mesenchymal stem cells promotes osteosarcoma development. Cell Death Dis. 2016;7(1):e2015. doi: 10.1038/cddis.2015.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bao Z., et al. Capsaicin induces cytotoxicity in human osteosarcoma MG63 cells through TRPV1-dependent and -independent pathways. Cell Cycle. 2019;18(12):1379–1392. doi: 10.1080/15384101.2019.1618119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X., et al. Wu X., et al. p38 MAPK regulates the expression of ether à go-go potassium channel in human osteosarcoma cells. Radiol. Oncol. 2013;47(1):42–49. doi: 10.2478/v10019-012-0043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu H.A. Prospective for the potential effect of local anesthetics on stem-like cells in colon cancer. Global J. Med. Res. 2020 https://medicalresearchjournal.org/index.php/GJMR/article/view/1971 [Review] [Google Scholar]
- 77.Liu H. A prospective for the role of two-pore channels in breast cancer cells. Global Journal of Cancer Therapy. 2020;6(1):1–3. [Google Scholar]