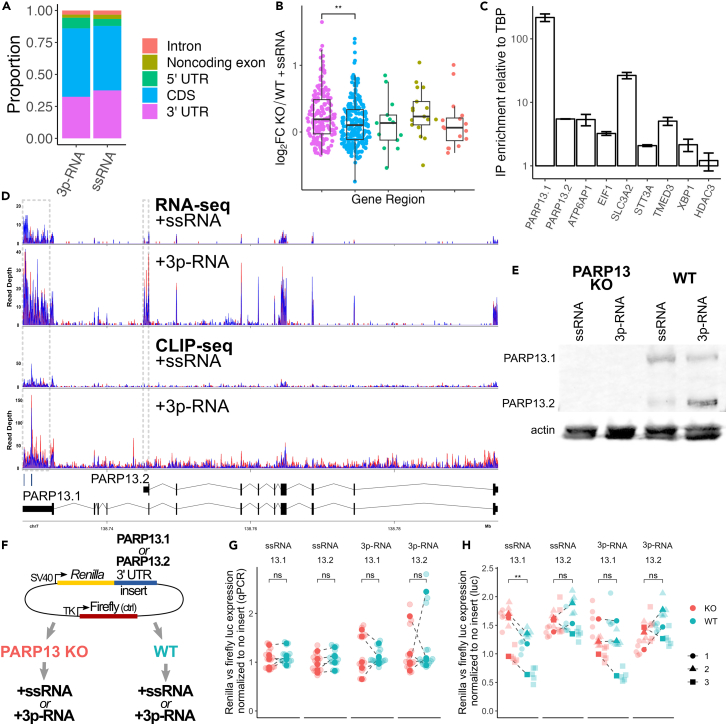
Figure 2.
PARP13 selectively binds PARP13.1 and regulates its translation via the 3′ UTR
(A) Proportions of PARP13 binding sites that fall within the indicated gene regions of mRNAs.
(B) Log2 fold-change RNA expression of PARP13-bound genes in PARP13 KO versus WT +ssRNA-treated cells, separated by what region of the transcript PARP13 binds. Transcripts where PARP13 binds multiple types of gene regions were excluded. Colors correspond to the gene region legend on the left. T-test, ∗∗ = p < 0.01.
(C) RNA immunoprecipitation using GFP-trap beads against exogenous GFP-PARP13.1 in HEK293T cells. Error bars are +/− 1sd.
(D) Aligned RNA-seq (top) and eCLIP-seq (middle) reads within PARP13 gene region for +ssRNA and +3p-RNA treatments with gray boxes surrounding the PARP13.1-specific (left) and PARP13.2-specific (right) 3′UTRs. Red and blue alignments correspond to duplicate experiments. Two PARP13 binding peaks located within the PARP13.1 3′UTR identified by CLIPper (Methods) are indicated below the aligned reads. Bottom: a splicing map of the two PARP13 isoforms, which are transcribed right to left.
(E) Western blot of PARP13 expression in WT and PARP13 KO HEK293T cell lysates 24 h after +ssRNA or +3p-RNA treatment, with beta-actin as a loading control. Also Figure S2A.
(F) Luciferase construct map and design of luciferase expression experiments.
(G) Transcript levels of Renilla luciferase with either PARP13.1 or PARP13.2 3′UTR treated with ssRNA or 3p-RNA (n = 5). Within each experiment, Renilla expression was normalized to firefly expression and compared to normalized Renilla expression for a construct without a 3′UTR insert. ns = not significant.
(H) Luminescence of Renilla luciferase (n = 3) with the same normalization as (G). Paired t-test, ∗∗ = p < 0.01; ns = not significant.