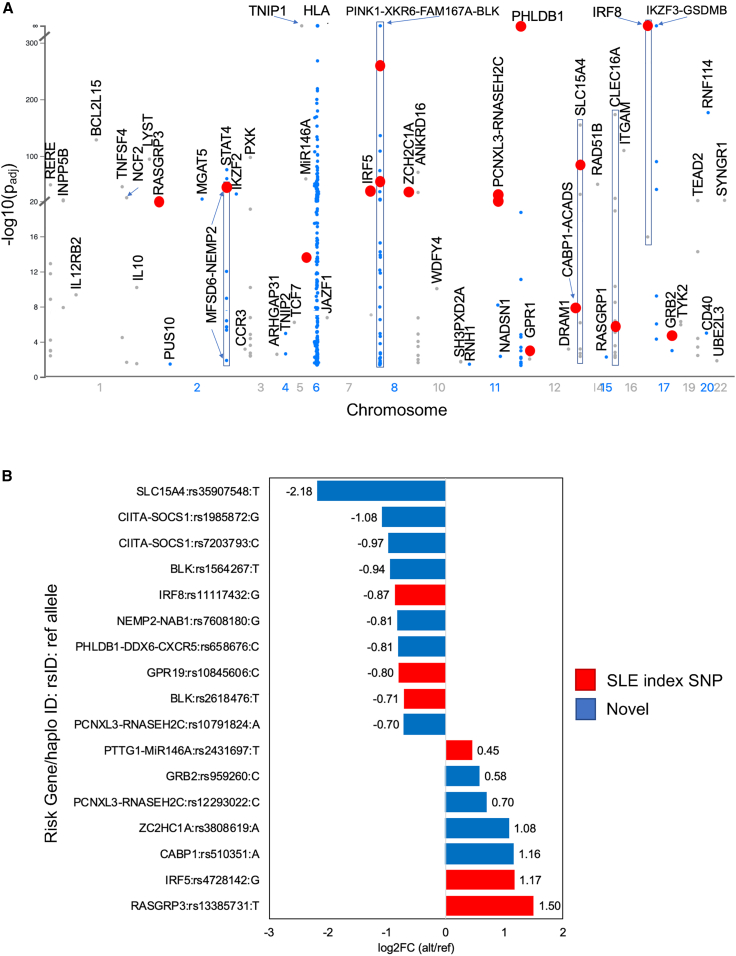
Figure 3.
EmSeq and emVar variants on SLE risk haplotypes
(A) Manhattan plot of significant emSeqs (−log10(padj) as determined by DESeq2 plotted on SLE risk haplotypes). Only the allele with the highest regulatory activity is plotted for each variant. SLE risk gene/haplotype is indicated. Red dots represent emVars.
(B) Effect sizes of emVars on SLE risk haplotypes. The risk gene/haplotype and reference allele are provided for each variant. Positive effects indicate the alternate allele significantly increases regulatory activity over the reference allele; negative effects indicate the reference allele significantly increases activity over the alternate allele. Candidate causal SLE index SNPs are indicated in red. Previously unreported candidate causal variants on SLE haplotypes are indicated in blue.