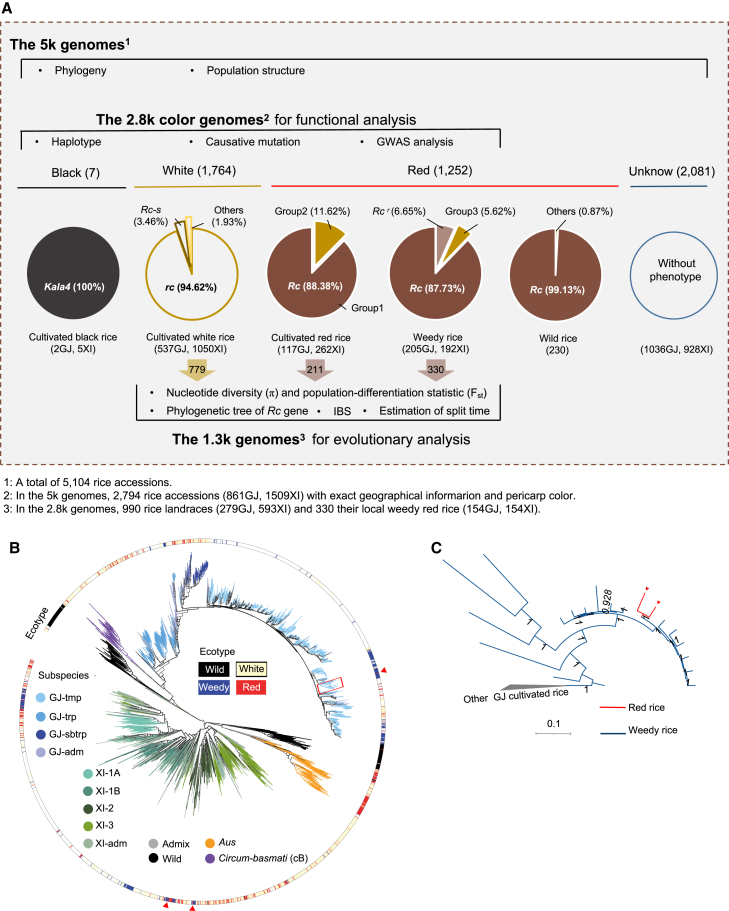
Figure 1.
Overview of the genomic data and workflow used in this study.
(A) Workflow used in this study. Among the 5k genomes, 2794 accessions with colored pericarps and geographical information (the 2.8k color genomes) were used for functional analysis, and 1320 rice accessions (the 1.3k genomes), including landraces and local weedy rice, selected from the 2.8k color genomes were used for evolutionary analysis. The accessions were classified into red, white, and black groups according to their available pericarp color. On the basis of genotypes of two functional genes (Rc and Kala4), the pigmented rice accessions (O. sativa) were further defined as black rice with the Kala4 genotype or red rice with the Rc/kala4 genotype. Most wild rice accessions (O. rufipogon) have red pericarps and the Rc/kala4 genotype.
(B) Phylogenetic tree of the 5k genomes inferred from whole-genome SNPs. Different subspecies are indicated by colored lines, and the outermost circle represents the ecotypes of the accessions.
(C) Detailed phylogenetic tree of the rice accessions in the red rectangle in (B), showing the relationship between a cultivated red rice (represented by the red lines) and weedy red rice. The cultivated red rice is surrounded by weedy red rice (blue lines). Other cultivated rice is represented by a gray cluster.