Figure 3.
Identification of new candidate genes/alleles for red pericarp in rice.
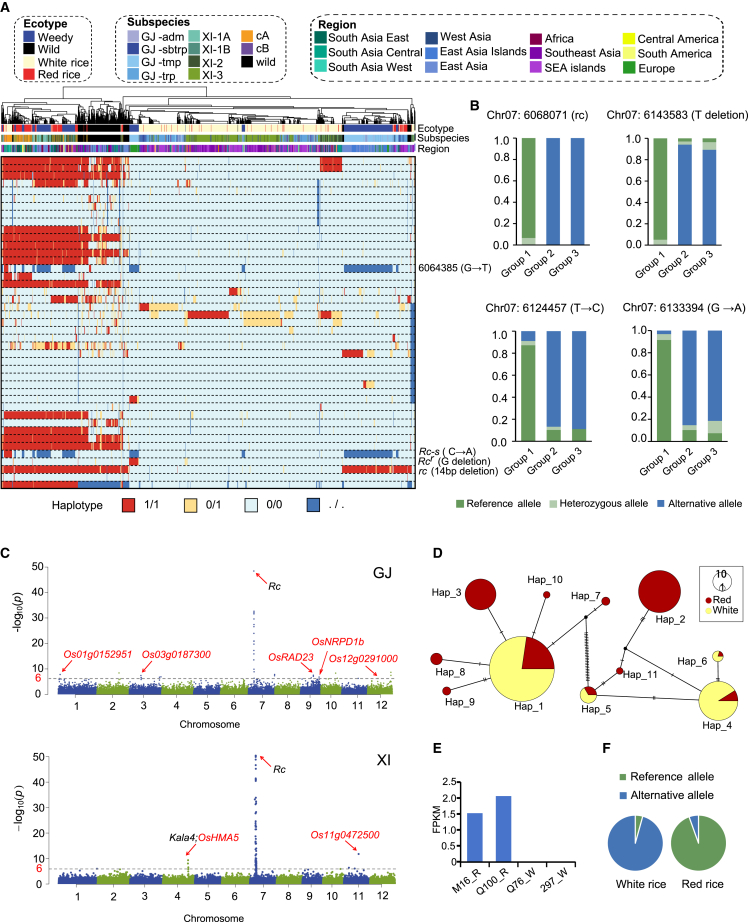
(A) SNP- and indel-based haplotype map of the Rc gene. The ecotype, subspecies, and source region of each accession are indicated by colored lines under the clustered tree, and important mutations, including rc, Rc-s, and Rcr, are marked at their positions. The genotypes (ref, alt, het, and missing) of each variant are color-coded, using the IRGSP-1.0 genome as the reference.
(B) Three possible sequence variants related to the contradiction between phenotype and genotype, including a deletion (T) and two single-base mutations (T to C and GnullA). All three variants had a higher rate of the reference allele in Group1 and a higher rate of the alternative allele in Group2 and Group3. Three groups were defined as Group#1 and Group#2, cultivated red rice with the Rc or rc allele, respectively, and Group#3, weedy red rice with the rc allele and without the Rcr gene.
(C) Manhattan plots for red pericarp color in the GJ (top) and XI (bottom) rice groups. The −log10(P) values from a genome-wide scan are plotted against position on each of 12 chromosomes. The horizontal dashed lines indicate the genome-wide significance thresholds (P = 2.9 × 10−7 for GJ and P = 3.0 × 10−7 for XI, equivalent to about 6.5 after the logarithm).
(D) Haplotype network of Os01g00152951 in the GJ group.
(E) Transcriptomic patterns of Os01g0152951 in the two red rice accessions presented as number of fragments per kilobase of exon model per million mapped reads (FPFM).
(F)Os04g0556000 in the XI group has a significantly different allele frequency between the red rice and white rice groups.