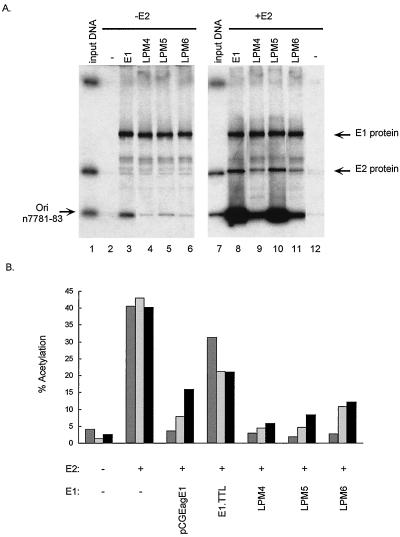
FIG. 4.
(A) DNA binding activity of E1 proteins containing mutations in the DNA binding domain. The left gel demonstrates E1 origin-specific binding, and the right gel demonstrates E1-E2 cooperative origin binding. Lanes 1 and 7 contain 1 ng (1/200) of input probe DNA, and the origin-containing fragment is indicated (Ori); lane 2 contains unprogrammed lysate; lanes 3 to 6 and 8 to 11 contain 35 μl of in vitro-translated E1 proteins, as described above each lane; and lanes 8 to 12 contain 25 μl of E2 protein lysate. The amount of total lysate per assay was kept constant by addition of control lysate. The positions of the E1 and E2 proteins are indicated. In each lane the DNA-protein complexes were immunoprecipitated with the E1-specific antibody SSQN (25), eluted from beads, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The percentages of origin binding were quantitated with a PhosphorImager, and levels of cooperative origin binding were expressed relative to the amount of binding found when only E1 was added, which was given a value of 1. The gel shows both 35S and 32P signals. n7781-83, nucleotides 7781 to 7783. (B) Effects of point mutations in the DNA binding domain of E1 on the protein’s ability to repress E2-mediated transactivation. The values were averaged from results of three experiments. Reporter plasmids p1066 (darkly shaded bars), p964 (lightly shaded bars), and pTKM6 (filled bars) were cotransfected into BEF cells with the indicated E1 (pCGEagE1) and/or E2 (C59) expression vector. Each plasmid was tested in several independent experiments. CAT activities are expressed as percentages of acetylation.