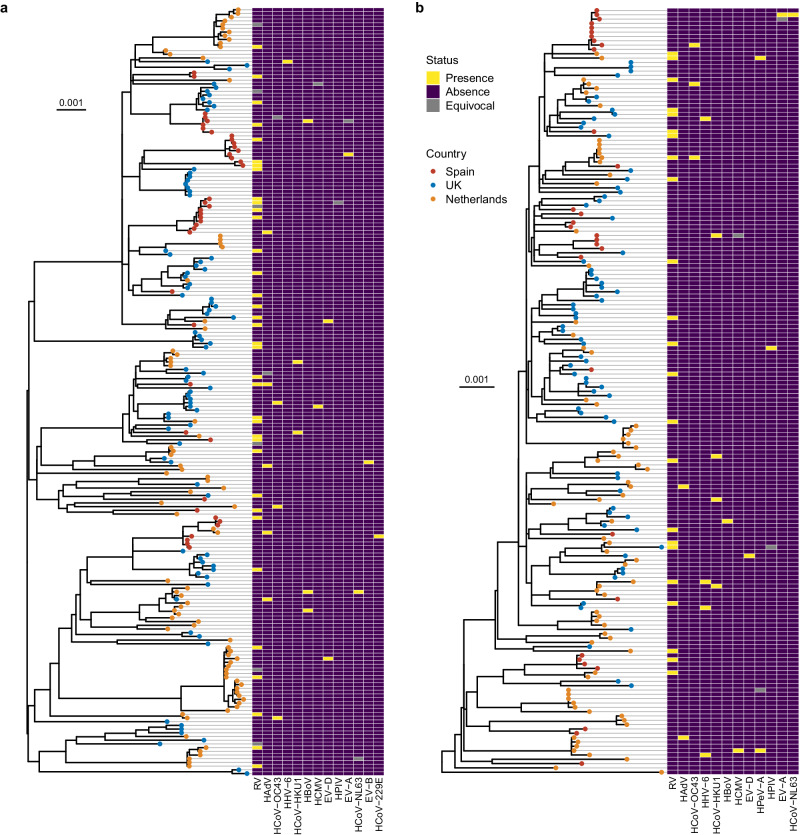
Fig. 3. Distribution of co-detected viruses on the RSV maximum-likelihood phylogenies.
a RSV-A phylogeny was reconstructed from 207 samples. b RSV-B phylogeny was reconstructed from 177 samples. At least 70% of the coding sequences were recovered from these samples. For infants with multiple samples collected, only the sample with the highest coverage was included. The trees were midpoint rooted. The sampling country of each strain is illustrated by tip colour. The scale bars represent the number of nucleotide substitutions per site. Human parechovirus A (HPeV-A) was not co-detected with any of the RSV-A strains; enterovirus B (EV-B) and human coronavirus 229E (HCoV-229E) were not co-detected with any of the RSV-B strains. Source data are provided as Source Data files. HAdV human adenovirus, HBoV human bocavirus, HCMV human cytomegalovirus, HHV-6 human herpesvirus 6, HPIV human parainfluenza virus, RSV respiratory syncytial virus, RV rhinovirus.