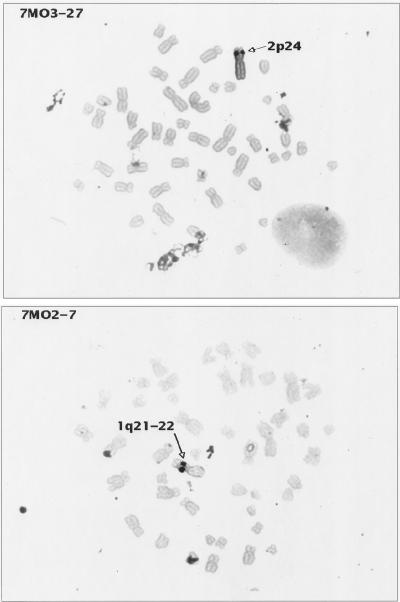
FIG. 4.
In situ hybridization of HTLV-2-transformed cell lines. Metaphase preparations of two HTLV-2 transformants (7MO2-7 and 7MO3-27) were hybridized with probe pMO-4. Cell cultures were harvested for metaphase preparation with colcemid by standard cytogenetic laboratory procedures. Probes were labelled by nick translation with either biotin-dUTP or digoxigenin-dUTP (Boehringer-Mannheim Corp.) and detected by nonfluorescent methods which use peroxidase-tagged avidin (Vector Lab Inc., Burlington, Calif.) or peroxidase-tagged anti-digoxigenin (Boehringer-Mannheim), followed by diaminobenzidine and silver amplification (Amersham Corp., Arlington Heights, Ill.). This method was originally described by Burns et al. (7), and hybridization and signal detection were performed as described by Lee et al. (36) with modifications (61). The probe concentration used was 10 μg/ml, and hybridization was performed overnight by standard in situ hybridization protocols (Oncor). For each probe and transformant cell line, several amplification times were tested: the best signal localization was achieved with the minimal amplification time (ranging from 20 to 40 min) sufficient to observe specific signal while minimizing the background. Cells were chosen for further analysis when the silver signals were confined to a single small dot on each chromatid. Images were recorded and analyzed with Cytovision software (Applied Imaging, Pittsburgh, Pa.). These results are representative of the results obtained with the other HTLV-2-transformed cell lines analyzed. Similar results were also achieved when HTLV-1-transformed cell lines were hybridized with the HTLV-1 probe pMT2, a full-length (8.25 kb) HTLV-1 probe as previously described (57).