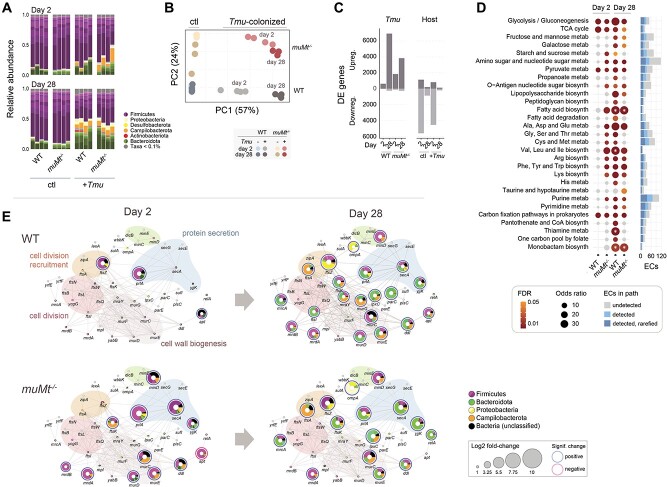
Figure 2.
Bacterial gene expression is altered in the presence of the protist. (A) Taxonomic profiles of putative bacterial mRNA reads. (B) Principal component analysis of bacterial gene expression. Circles denote samples isolated from individual mice, colored by host B-cell status, presence of Tmu and timepoint, as shown. Expression values are based on DESeq2-variance stabilized counts. (C) Numbers of significantly upregulated or downregulated bacterial genes between protist-colonized and naïve mice, or due to host B-cell status, grouped as indicated (n = 3 or 4). (D) Bacterial metabolic pathway enrichment associated with Tmu colonization. Enrichment was determined by the overrepresentation of EC terms in KEGG-defined pathways within DE genes. Total ECs detected in the experiment and in the rarefied gene expression matrix used in the analysis are indicated in the bar chart to the right. Significantly enriched pathways are represented as colored dots: gray indicates ns, and the gradient from yellow to red represents decreasing P values, beginning from P < .05. Sizes of dots represent odds ratios. Significance was calculated using Fisher’s exact tests. Asterisks indicate infinite odds ratios. (E) Network of cell division, cell wall biogenesis, and secretory genes, based on a previously determined E. coli protein network [52]. Colored nodes represent protein homologs identified in the metatranscriptomic data and edges are previously described protein–protein or functional interactions. Nodes in gray represent undetected genes. Genes significantly up- and downregulated are denoted with blue and red borders, respectively. Node sizes correlate with log2 fold-changes between Tmu-colonized and naïve mice. Colors within node pie charts indicate proportions of gene expression assigned to various taxa, as shown. Colors encircling groups of proteins define functional modules.