Abstract
The gypsy element of Drosophila melanogaster is the first retrovirus identified so far in invertebrates. Previous data suggest that gypsy ENV-like ORF3 mediates viral infectivity. We have produced in the 293GP/LNhsp70lucL.3 human cell line a Moloney murine leukemia virus-based retroviral vector pseudotyped by the gypsy ENV-like protein. We have shown by immunostaining that the gypsy envelope protein is produced in 293GP/LNhsp70lucL.3 cells and that vector particles collected from these cells can infect Drosophila cells. Our results provide direct evidence that the infectious property of gypsy is due to its ORF3 gene product.
The gypsy (or mdg-4) element is a 7,469-bp retroelement (11) that resides within the genome of Drosophila melanogaster. gypsy shares several structural and functional properties with the vertebrate retroviruses (reviewed in reference 14): (i) gypsy is flanked by long terminal repeats (LTRs) with a U3-R-U5 retroviral structure that directs transcription, (ii) gypsy has three open reading frames (ORFs) analogous in size and position to the retroviral gag, pol, and env genes (Fig. 1), and (iii) the organization of the pol gene is similar to that of the vertebrate retroviral pol genes (Fig. 1).
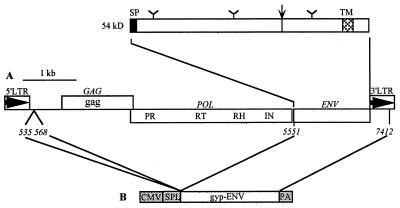
FIG. 1.
(A) Organization of the gypsy element. Two LTRs flank a central region which contains the three ORFs corresponding to gag, pol, and env. The order of the various domains of the pol gene is indicated (PR, protease; RT, reverse transcriptase; RH, RNase H; IN, integrase). The predicted 54-kDa protein encoded by ORF3 is represented with potential N-glycosylation sites (Y), the signal peptide (SP), the dibasic cleavage site (arrow), and the transmembrane domain (TM). (B) Genetic organization of pHCMV-gyp-ENV. Numbers refer to gypsy nucleotides. CMV, human cytomegalovirus immediate-early promoter; SPL and PA, splicing and polyadenylation signals, respectively, derived from the rabbit beta-globin gene.
The presence of a third ORF is characteristic of the invertebrate gypsy family retroelements, including among others 17.6 (16), 297 (7), and tom in Drosophila (18) and TED in Trichoplusia (10). The predicted 54-kDa protein encoded by gypsy ORF3 shares several general features with vertebrate retroviral ENV glycoproteins (13, 17) (Fig. 1). It contains a potential signal peptidase cleavage site at its N terminus and a potential protease cleavage site Arg-Ser-Arg-Arg similar to the vertebrate retroviral ENV cleavage motif (Arg/Lys)-X-Lys-Arg. There are three consensus N-linked glycosylation sites and a C-terminal transmembrane domain.
gypsy is controlled by the flamenco gene of D. melanogaster (15). The gypsy ORF3 RNA and protein are detected only in females homozygous for flamenco permissive alleles (13, 17). It has been shown that extracts of homozygous flamenco permissive pupae and females can infect flamenco permissive larvae (9, 17) and that these female extracts contain infectious gypsy associated with the product of gypsy ORF3 (17). These data support the concept that gypsy is an autonomous retrovirus, the first identified so far in invertebrates, and that gypsy ENV-like ORF3 mediates viral infectivity. To analyze the infectious properties of gypsy ENV-like ORF3, we packaged a mammalian retroviral vector based on Moloney murine leukemia virus (MoMLV) in the gypsy ENV-like protein.
MoMLV-based retroviral vectors can be pseudotyped with gypsy envelope.
Previous studies have shown that the MoMLV recombinant genome can be pseudotyped with the vesicular stomatitis virus envelope glycoprotein (VSV-G) (2). To test whether the MoMLV genome could be pseudotyped with the gypsy envelope protein (gyp-ENV), the human cell line 293GP/LNhsp70lucL.3 (LNhsp70lucL) (8) (Fig. 2) was transiently transfected with the pHCMV-gyp-ENV plasmid expressing the gypsy ORF3 under the control of the strong immediate-early promoter of human cytomegalovirus. To construct pHCMV-gyp-ENV, a BamHI-BglII fragment containing a part of the leader region (nucleotides 535 to 568; accession no. M12927 [11]), ORF3, and a part of the 3′ LTR (nucleotides 5551 to 7412) of gypsy were inserted into the unique BamHI site in pHCMV-Bam (19).
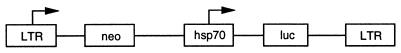
FIG. 2.
Genetic organization of LNhsp70lucL provirus. LTR, MoMLV LTR; neo, neomycin phosphotransferase gene; hsp70, D. melanogaster heat shock 70 promoter; luc, firefly luciferase gene. Arrows indicate direction of transcription.
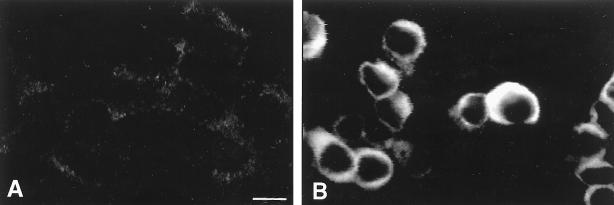
The expression of gypsy envelope protein in LNhsp70lucL cells was assessed by immunostaining with the 7B3 monoclonal antibody (Ab) raised against the gypsy ORF3 product (17). Seventy-two hours after transfection with pHCMV-gyp-ENV, cells were washed in phosphate-buffered saline (PBS), fixed in cold acetone, washed in PBS, and then incubated sequentially with the 7B3 monoclonal Ab (diluted 1/2) for 1 h and fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (Vector Laboratories) (diluted 1/100) for 40 min. The expression of gypsy envelope protein was identified in transfected LNhsp70lucL cells by confocal microscopy (Fig. 3).
FIG. 3.
Localization of the gypsy envelope protein by confocal microscopy. (A) Negative control: nontransfected LNhsp70lucL cells stained by anti-gyp-ENV and anti-mouse FITC antibodies; (B) LNhsp70lucL cells transiently transfected with pHCMV-gyp-ENV and stained with both anti-gyp-ENV and anti-mouse FITC antibodies. Bar, 10 μm.
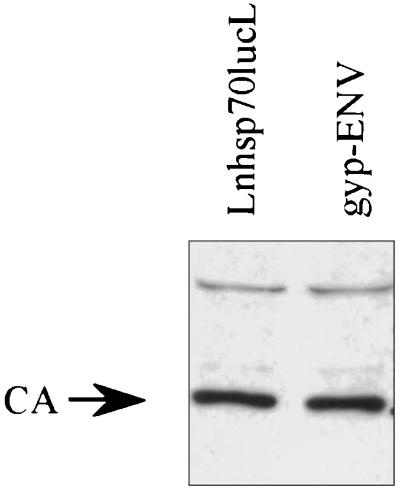
The infection efficiency of the retroviral vector packaged with gyp-ENV was determined and compared to that observed with the same retroviral vector packaged with the VSV-G. These VSV-G-pseudotyped vectors have previously been shown to infect and stably integrate into insect cells (12). Briefly, 106 LNhsp70lucL cells were plated onto 100-mm-diameter plates overnight. Cells were transfected with 20 μg of the appropriate plasmid by standard calcium phosphate coprecipitation. At 72 h posttransfection, cell culture supernatants were collected, filtered (pore size, 0.45 μm), and diluted 1/2 in Schneider medium (GIBCO) supplemented with 10% fetal bovine serum and 2-μg/ml Polybrene, a polycation that increases infection efficiency. Pseudotyped vectors were produced as previously described (19). Supernatants harvested from LNhsp70lucL cells with no envelope protein contain noninfectious viral particles and were used as a negative control. A Western blot was performed in order to compare the relative number of particles produced by gyp-ENV-expressing producer cells and producer cells with no envelope protein. Virus supernatants were concentrated by ultracentrifugation (SW41 rotor, 30,000 rpm for 1 h at 4°C), and equivalent volumes of resuspended viral pellets were loaded on a 10% polyacrylamide gel. Immunostaining for the MoMLV viral core protein with anti-p30 antibody was performed as previously described (3) and revealed that approximately equal numbers of viral particles were present in each viral pellet (Fig. 4). Thus, the noninfectious viral supernatant and the supernatant containing the pseudotyped vector contain approximately equal numbers of viral particles.
FIG. 4.
Immunoblot of pellets of viral particles produced from gyp-ENV-expressing cells and LNhsp70lucL cells with no envelope protein. The blot was stained with anti-p30 antiserum to detect the CA capsid protein.
The Schneider 2 D. melanogaster somatic cells were used as recipient cells for the infection experiments. These cells do not express the gypsy envelope as determined by immunostaining with the 7B3 Ab (data not shown). Subconfluent Schneider 2 cells were incubated with these supernatants. Virus-containing supernatant was replaced by fresh Schneider medium 24 h after infection. Centrifugation, at 1,000 × g for 1 h, of cells immediately after the addition of the supernatant was tested as a means to increase infection efficiency (Table 1). The influence of gravitational force during centrifugation on the net direction of Brownian movement of virus particles can increase the probability of contact between the virus and the cell surface (1).
TABLE 1.
Luciferase activity in Schneider 2 cells infected by retroviral vector LNhsp70lucL pseudotyped by gyp-ENV or VSV-G envelope
| Envelope expression | Light units/μg of proteina
|
||
|---|---|---|---|
| No centrifugation
|
Centrifugation,b no heat shock | ||
| No heat shock | Heat shock | ||
| Transient | |||
| No envelope | 0.7d | NDc | 0.7d |
| gyp-ENV | 1.4 ± 0.4 | ND | 19.7 ± 12.3 |
| VSV-G | 998.0 ± 223.0 | ND | 2352.0 ± 148.5 |
| Established | |||
| No envelope | 6.7d | 3.5d | 3.8d |
| gyp-ENVe | 20.8 ± 10.9, 21.0 ± 12.3 | 40.3 ± 14.7, 31.6 ± 4.4 | 243.7 ± 111.6, 255.3 ± 67.5 |
The data are means of three assays ± standard deviations except as indicated.
Cells in a tissue culture well plus virus were centrifuged at 1,000 × g for 1 h.
ND, not determined.
Single assay.
Results obtained with two different supernatants in two separate experiments.
To test for evidence of infection, cells were washed 4 days postinfection in PBS, lysed, and briefly centrifuged, and supernatants were assayed for luciferase activity according to the manufacturer’s (Promega) instructions. Infection with the gyp-ENV-pseudotyped MoMLV was clearly demonstrated following centrifugation (Table 1). The luciferase activity measured after infection with the VSV-G-pseudotyped MoMLV was at least 100-fold higher than the activity measured after infection with the gyp-ENV-pseudotyped MoMLV.
To examine the host range of the gyp-ENV-pseudotyped particles and to confirm the persistence of proviral DNA, MOS-55 cells from the mosquito Anopheles gambiae were infected with supernatant containing noninfectious viral particles or gyp-ENV-pseudotyped particles. Infections were performed in triplicate, cells were harvested at 72 h postinfection, and cell lysates and DNA were prepared as previously described (7). No detectable luciferase activity was observed in MOS-55 cells infected with or without centrifugation (mean light units per microgram of protein: no centrifugation, 1.3 ± 0.1; with centrifugation, 1.4 ± 0.3; negative control, 1.0). Amplification with nested primers specific for the proviral LTR (7) yielded the expected 176-bp amplicon from the mosquito cell lines infected with the gyp-ENV-pseudotyped vector. No PCR product was seen following amplification of DNA from mock-infected cells, despite a positive signal following amplification with muscle actin primers (7). These results suggest that the gyp-ENV-pseudotyped particles are weakly infectious in MOS-55 cells.
Hence, our results demonstrate that the gyp-ENV-pseudotyped vector was able to infect Schneider 2 cells, although the infection efficiency was lower than that for the VSV-G-pseudotyped vectors.
gypsy envelope protein can be stably expressed in LNhsp70lucL packaging cell lines.
Because expression of gypsy envelope did not appear to be toxic in LNhsp70lucL cells (no syncytia were observed), we generated a stable cell line that constitutively expresses the gypsy envelope. Cells were cotransfected with a plasmid conferring resistance to puromycin and pHCMV-gyp-ENV, at a ratio of 1:5. Transfected cells were selected in puromycin (1 μg/ml) and G418 (400 μg/ml). Colonies were screened for their ability to produce infectious pseudotypes by incubating culture supernatants with Schneider 2 cells and assaying for luciferase activity 4 days postinfection. The clone producing the highest virus titer was chosen, and expression of gyp-ENV was verified by staining with anti-gyp-ENV.
To increase the level of luciferase expression, infected Schneider 2 cells were heat shocked (30 min at 37°C, 1 h of recovery at 23°C, 30 min at 37°C, and 45 min at 23°C) prior to analysis of luciferase activity. The effects of different concentrations of Polybrene on infection efficiency were tested, and maximal luciferase activities were obtained with 8 μg/ml.
The luciferase activity observed following infection with culture supernatants derived from the packaging cell lines constitutively expressing the gypsy envelope was markedly increased compared to infection with supernatants produced after transient transfection (Table 1). As expected, heat shock activated the hsp70 promoter, thus increasing luciferase activity.
In this study, we show for the first time that the vertebrate retroviral recombinant genome MoMLV can be pseudotyped by the invertebrate retroviral gypsy envelope protein. Our results provide direct evidence that the infectious properties of gypsy (9, 17) are due to its ORF3 gene product.
The fact that no cell fusion was observed in the LNhsp70lucL cell line constitutively expressing the gyp-ENV protein may be due to the absence of an appropriate receptor on the mammalian cell surface or to a low abundance of the gypsy ENV protein expressed in these cells (6).
Our data suggest that the gypsy envelope is correctly processed in human cells. Transport of the retroviral envelope protein to the cell surface and its proteolytic cleavage are necessary steps for incorporation into virus-like particles and budding of infectious particles. Among the endoproteases, it has been shown that the furin protease has a role in cleavage of the human immunodeficiency virus envelope proteins (5). Dfur1 and Dfur2 (4) are two D. melanogaster genes exhibiting homology with the human furin. This suggests that furin-like function is conserved among taxa, which could explain the faithful processing of the gypsy envelope protein in mammalian cells.
Retroviral pseudotypes produced from our stable packaging cell line expressing the gypsy envelope protein provide a convenient method to analyze the tropism of the gypsy envelope in different insect species. The gypsy envelope-pseudotyped vector as a transformation vector in invertebrates other than D. melanogaster is under investigation in our lab.
Acknowledgments
We are grateful to Alain Pélisson for helpful discussions and critical reading of the manuscript. We thank Fabienne Chalvet for providing the gypsy fragment containing the ORF3 gene. We thank Bernard Mignotte’s group for help with cell culture techniques, Spencer Brown for help with confocal microscopy, and Jean-Pierre Rousset for help with the luciferase activity determination. We thank François-Loïc Cosset for helpful discussion and the p30 anticapsid Ab. The 7B3 Ab was kindly provided by Victor Corces, Baltimore, Md.
L.T. is a recipient of a fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche. This work was supported by grants from the Centre National de la Recherche Scientifique, the Association pour la Recherche sur le Cancer (contract 1132), and the Actions Coordonnées Concertées des Sciences du Vivant.
REFERENCES
- 1.Andreadis S T, Palsson B O. Kinetics of retrovirus mediated gene transfer: the importance of intracellular half-life of retroviruses. J Theor Biol. 1996;182:1–20. doi: 10.1006/jtbi.1996.0140. [DOI] [PubMed] [Google Scholar]
- 2.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosset F L, Morling F J, Takeuchi Y, Weiss R A, Collins M K, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Bie I, Savaria D, Roebroek A J, Day R, Lazure C, Van de Ven W J, Seidah N G. Processing specificity and biosynthesis of the Drosophila melanogaster convertases dfurin1, dfurin1-CRR, dfurin1-X, and dfurin2. J Biol Chem. 1995;270:1020–1028. doi: 10.1074/jbc.270.3.1020. [DOI] [PubMed] [Google Scholar]
- 5.Decroly E, Vandenbranden M, Ruysschaert J M, Cogniaux J, Jacob G S, Howard S C, Marshall G, Kompelli A, Basak A, Jean F, et al. The convertases furin and PC1 can both cleave the human immunodeficiency virus (HIV)-1 envelope glycoprotein gp160 into gp120 (HIV-1 SU) and gp41 (HIV-I TM) J Biol Chem. 1994;269:12240–12247. [PubMed] [Google Scholar]
- 6.Gething M J, Doms R W, York D, White J. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J Cell Biol. 1986;102:11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inouye S, Yuki S, Saigo K. Complete nucleotide sequence and genome organization of a Drosophila transposable genetic element, 297. Eur J Biochem. 1986;154:417–425. doi: 10.1111/j.1432-1033.1986.tb09414.x. [DOI] [PubMed] [Google Scholar]
- 8.Jordan, T. V., H. Shike, V. Boulo, V. Cedeno, Q. Fang, B. S. Davis, M. Jacobs-Lorena, S. Higgs, K. J. Fryxell, and J. C. Burns. Pantropic retroviral vectors mediate efficient cellular transformation and expression of foreign genes in insects. Insect. Mol. Biol., in press. [DOI] [PubMed]
- 9.Kim A, Terzian C, Santamaria P, Pelisson A, Prud’homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lerch R A, Friesen P D. The baculovirus-integrated retrotransposon TED encodes gag and pol proteins that assemble into viruslike particles with reverse transcriptase. J Virol. 1992;66:1590–1601. doi: 10.1128/jvi.66.3.1590-1601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marlor R L, Parkhurst S M, Corces V G. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol. 1986;6:1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsubara T, Beeman R W, Shike H, Besansky N J, Mukabayire O, Higgs S, James A A, Burns J C. Pantropic retroviral vectors integrate and express in cells of the malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci USA. 1996;93:6181–6185. doi: 10.1073/pnas.93.12.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelisson A, Song S U, Prud’homme N, Smith P A, Bucheton A, Corces V G. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelisson, A., L. Teysset, F. Chalvet, A. Kim, N. Prud’homme, C. Terzian, and A. Bucheton. About the origin of retroviruses and co-evolution of the gypsy retrovirus with the Drosophila flamenco host gene. Genetica, in press. [PubMed]
- 15.Prud’homme N, Gans M, Masson M, Terzian C, Bucheton A. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saigo K, Kugimiya W, Matsuo Y, Inouye S, Yoshioka K, Yuki S. Identification of the coding sequence for a reverse transcriptase-like enzyme in a transposable genetic element in Drosophila melanogaster. Nature. 1984;312:659–661. doi: 10.1038/312659a0. [DOI] [PubMed] [Google Scholar]
- 17.Song S U, Gerasimova T, Kurkulos M, Boeke J D, Corces V G. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 18.Tanda S, Mullor J L, Corces V G. The Drosophila tom retrotransposon encodes an envelope protein. Mol Cell Biol. 1994;14:5392–5401. doi: 10.1128/mcb.14.8.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yee J K, Miyanohara A, LaPorte P, Bouic K, Burns J C, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]